Fig. 7.
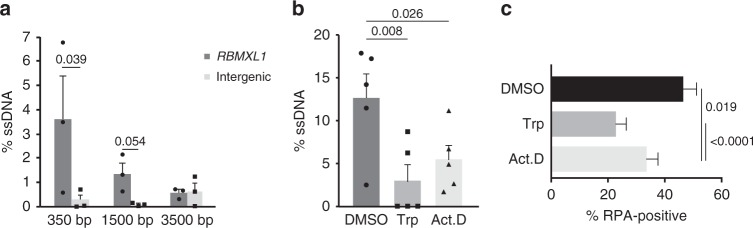
Transcription facilitates DNA end resection. a DIvA cells were incubated for 4 h with 300 nM 4-OHT and DNA end resection was measured by qPCR at three positions upstream of an AsiSI cleavage site located in a silent, intergenic sequence. The plot compares the percentage of ssDNA at the intergenic locus with that calculated for the transcribed RBMXL1 locus in Fig. 3c. The error bars represent s.e.m. and statistical testing was done using a two-tailed Student’s t-test (n = 3 independent experiments). b DNA end resection at the RBMXL1 locus was also quantified in DIvA cells treated with either 10 µM Triptolide for 30 min or 8 µM ActD for 1 h. The plot shows the percentage of ssDNA 350 nt upstream of the AsiSI cleavage site in control cells (DMSO) and cells treated with RNAPII inbibitors. The error bars represent s.e.m. and statistical testing was done using a one-tailed Student’s t-test (n = 5 independent experiments). c The effect of transcription inhibition on the formation of RPA foci was analysed in HeLa cells exposed to ionizing radiation (5 Gy) and stained with antibodies against RPA and γ-H2AX 15 min after irradiation. The bar plot shows the percentage of cells with γH2AX-positive foci that were co-stained by RPA. Error bars show s.e.m. Statistical testing was done using a Mann–Whitney’s test (n > 100 cells analysed in each condition, from four independent experiments). Source data for Fig. 7a–c are provided as a Source Data file