Figure 6. Model Prediction and Validation for a Dynamic Perturbation of Aging.
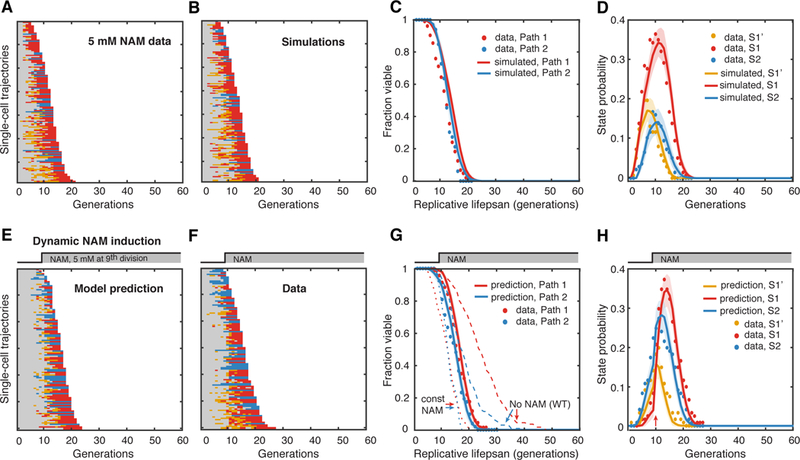
(A) Single-cell state transition trajectories under 5 mM constant NAM from the data (137 cells with 104 in Path 1, 33 in Path 2) and (B) from stochastic simulations. (C) The lifespans of two aging paths under 5 mM constant NAM from experimental data (red and blue solid circles) and from simulations (red and blue curves). (D) Age-dependent state distributions of S1’, S1, S2 under 5 mM NAM. Solid circles represent the experimental data. Solid curves represent simulated results averaged from 50 simulations with shaded areas indicating standard deviation. (E) Model-predicted single-cell state transition trajectories of WT cells in response to the step input of 5 mM NAM at 9th generation. (F) Experimental state transition trajectories of WT cells with the step input of 5 mM NAM after 600 mins, when more than 70% of cells just enter their 9th generation. (G) The lifespans of two aging paths in response to the dynamic perturbation. Predictions: red and blue curves; Experimental data: red and blue solid circles. Dashed curves are the lifespans for no NAM treatment and constant NAM treatment. (H) Model-predicted age-dependent state distributions of S1’, S1, S2. Note the sharp increase of S1 and S2 at 10th division (pointed by the red arrow). Solid curves are predictions averaged from 50 simulations with shaded areas indicating standard deviation. Solid circles represent the experimental data. See also Table S1.
