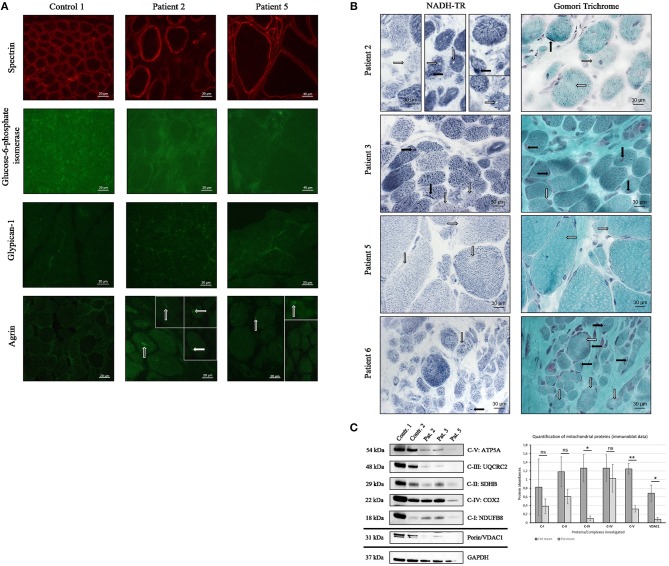
Figure 4.
Confirmation study of proteomic findings in independent MDC1A-patient derived muscle biopsies. (A) Immunofluorescence studies of spectrin have been carried out to visualize sarcolemma in muscle cryostat sections of control and patient-derived muscles and to show similar abundance and localization of the protein. Further immunofluorescence studies of glucose-6-phosphate isomerase reveal a focal dot-like sarcoplasmic staining/ enrichment in muscle biopsy specimen derived of controls (one representative control is shown here) but show a decrease in the biopsies of two representative MDC1A-patients. Immunofluorescence studies of glypican-1 showed focal enrichment at the sarcolemma of control muscle (one representative control is shown). However, in the LAMA2-diseased muscle fibers, small dots sarcoplasmic, and (sub-)sarcolemmal dots immunoreactive for glypican-1 were detected. Immunofluorescence studies of agrin revealed of a sarcoplasmic enrichment/ aggregation in MDC1A-patient derived muscle fibers (white arrows) compared to control muscle fibers. (B) Histological NADH-TR staining revealed “sarcoplasmic gaps” (white arrows in patient 2; second and third column) and reduced enzyme activity (white arrows in patient 2, 3, 5, and 6) as well as focal enrichment in some myofibers (black arrows in patient 2, 3, and 6). Modified Gomori Trichrome staining revealed sarcoplasmic areas with minor staining (white arrows in patient 2, 3, 5, and 6) as well as areas of sarcoplasmic and subsarcolemmal enrichment (black arrows in patient 2,3, and 5). (C) Immunoblot analysis of proteins representative for the respiratory chain complexes I-V and of VDAC1 (porin) revealed a statistically significant decrease of complexes III and V as well as of VDAC1 in MDC1A-patient derived protein muscle extracts compared to controls (diagram). GAPDH has been used as loading control and proteins have been quantified against GAPDH (ns, not significant; *statistically significant, **statistically very significant).