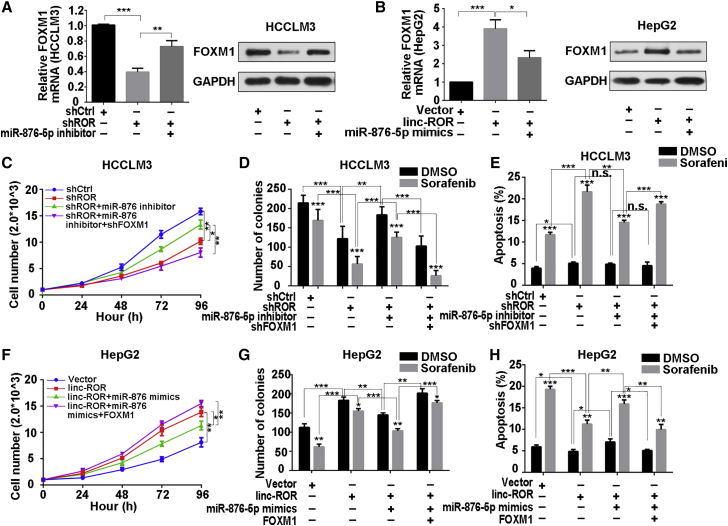
Figure 5.
FOXM1 Suppression, Mediated by miR-876-5p, Was Involved with LINC-ROR-Mediated Sorafenib Tolerance
(A) Quantitative real-time PCR and (B) western blotting were utilized to examine FOXM1 mRNA and protein levels in HCCLM3 cells, transfected with shROR or co-transfected with shROR and miR-876-5p inhibitor, or in HepG2 cells, transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics. (C) CCK-8 assays were carried out to detect the proliferation of HCCLM3 cells transfected with shROR or co-transfected with shROR and miR-876-5p inhibitor or co-transfected with shROR, miR-876-5p inhibitor, and shFOXM1. (D) Quantification of colony-formation ability of HCCLM3 cells transfected with shROR or co-transfected with shROR and miR-876-5p inhibitor or co-transfected with shROR, miR-876-5p inhibitor, and shFOXM1 under sorafenib treatment. (E) Quantification of total apoptosis rate of HCCLM3 cells treated with shROR or co-transfected with shROR and miR-876-5p inhibitor or co-transfected with shROR, miR-876-5p inhibitor, and shFOXM1, with the addition of DMSO or sorafenib. The percentage represents a sum of the Annexin- and 7-AAD-positive populations. (F) CCK-8 assays were carried out to detect the proliferation of HepG2 cells transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics or co-transfected with LINC-ROR, miR-876-5p mimics, and FOXM1. (G) Quantification of colony-formation ability of HepG2 cells transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics or co-transfected with LINC-ROR, miR-876-5p mimics, and FOXM1 after sorafenib treatment. (H) Quantification of total apoptosis rate of HepG2 cells transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics or co-transfected with LINC-ROR, miR-876-5p mimics, and FOXM1, with the addition of DMSO or sorafenib. The percentage represents the sum of the Annexin- and 7-AAD-positive populations. All data are presented as the mean ± SD of three independent experiments. The p values represent comparisons between groups (*p < 0.05, **p < 0.01, ***p < 0.001).