Figure 7.
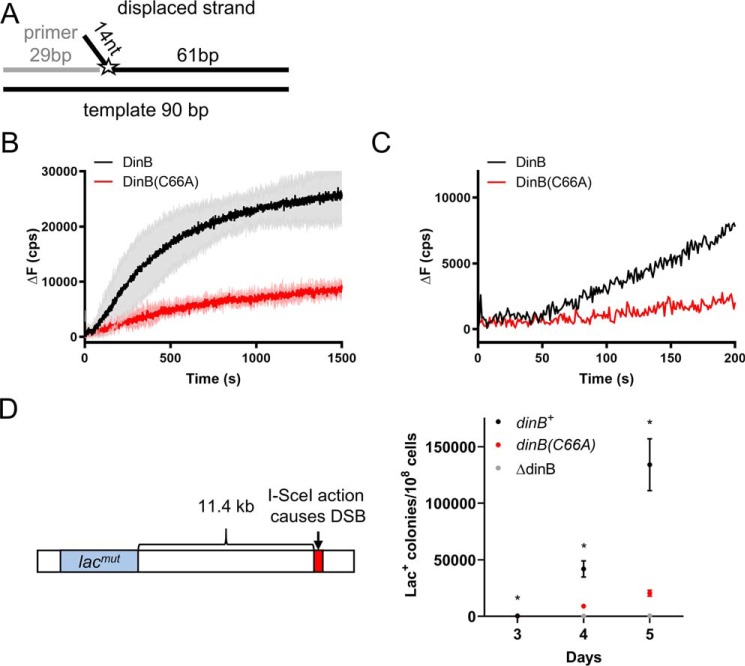
DinB(C66A) variant is deficient in RecA-independent strand displacement. A, graphic depiction of the DNA substrate used in these experiments. A 29-nt ssDNA primer (gray line) was annealed to a 90-nt template (bottom black line) as well as to a 75-nt fluorescently-labeled oligonucleotide (top black line with the fluorophore represented by the star). The 75-nt oligonucleotide displaced strand is composed of a 61-nt complementary to the 90-bp template and of a 14-nt unannealed flap located at the 5′ end. The fluorescein label (depicted by the star) on the displaced strand was located on the first nucleotide of the 75-bp complementary region. If displaced and template strands are separated, fluorescence is altered. DinB must insert a single nucleotide onto the end of the primer to displace the labeled nucleotide. B, experiments with all dNTPs added show that DinB stabilizes strand displacement after a short lag (highlighted by the enlargement in C) with greater efficiency than the DinB(C66A) variant. Initial velocities for both proteins from 0 to 50 s are not significantly different from zero. The rate of the DinB reaction from 100 to 200 s (46.40 ± 1.188 cps) is significantly higher than the velocity of the DinB(C66A) reaction at the same time point (11.80 ± 1.140 cps; p value < 0.0001). The DinB reaction also reaches a significantly higher maximum ΔF (28,053 ± 6272 cps) than the DinB(C66A) reaction (9798 ± 1290 cps; p value < 0.01). Maximum ΔF was measured using a measure of the fluorescence caused by the outgoing strand alone at about 37,000 cps. Change in fluorescence was measured in counts/s with respect to the fluorescence at 0 s. Experiments were performed in triplicate. Mean ± S.D. (shaded region surrounding curves) is shown in B. Only the mean is shown in C for better visualization of lag. D, E. coli strain containing a system described by Ponder et al. (31) was used to examine mutagenesis in response to an induced DSB (left). The endonuclease I–SceI created a DSB 11.4 kb on the E. coli chromosome downstream of the lac gene. The lac gene in this strain contains a mutation preventing cell growth when the only carbon source is lactose represented here by lacmut. The Lac− to Lac+ reversion is measured as the number of Lac+ colonies observed when the test strain is grown on lactose-only containing minimal medium divided by the total number of colonies observed when the strain is grown on glucose-only containing minimal medium (right). The (C66A) mutation in DinB (red) significantly reduces the rate of Lac+ reversion in the presence of a DSB. For comparison purposes, we used ΔdinB and dinB+ strains shown in gray and black, respectively. Data shown is mean of 12 replicates ± S.E. * indicates a p value < 0.05 when comparing the dinB+ and dinB(C66A) strains by two-tailed t test.