Abstract
Platelet factor 4 (PF4) is an anti-Plasmodium component of platelets. It is expressed in megakaryocytes and released from platelets following infection with Plasmodium. Innate immunity is crucial for the host anti-Plasmodium response, in which type I interferon plays an important role. Whether there is cross-talk between innate immune signaling and the production of anti-Plasmodium defense peptides is unknown. Here we demonstrate that E74, like ETS transcription factor 4 (ELF4), a type I interferon activator, can help protect the host from Plasmodium yoelii infection. Mechanically, ELF4 binds to the promoter of genes of two C-X-C chemokines, Pf4 and pro-platelet basic protein (Ppbp), initiating the transcription of these two genes, thereby enhancing PF4-mediated killing of parasites from infected erythrocytes. Elf4−/− mice are much more susceptible to Plasmodium infection than WT littermates. The expression level of Pf4 and Ppbp in megakaryocytes from Elf4−/− mice is much lower than in those from control animals, resulting in increased parasitemia. In conclusion, our study uncovered a distinct role of ELF4, an innate immune molecule, in host defense against malaria.
Keywords: malaria, mouse, Plasmodium, innate immunity, transcription regulation, ELF4, PF4, PPBP
Introduction
Malaria is a major public health problem, especially in Africa and Southeast Asia (1). Drug resistance of Plasmodium parasites remains a pressing concern (2). Nowadays, artemisinin combination therapy, which contains an artemisinin derivative, is the standard treatment for Plasmodium falciparum infection, according to guidelines for the treatment of malaria (3). However, artemisinin-resistant parasites have been found in western Cambodia (4, 5). There is a great need for new and effective approaches to fight malaria.
The innate immune system acts as the first defense against pathogens, relying on recognition of pathogen-associated molecular patterns by various kinds of pattern recognition receptors (PRRs)5 (6, 7). Well-known PRRs include Toll-like receptors (8), RIG-I-like (retinoic acid-inducible gene I-like) receptors (9), and several nucleic acid sensors, such as cGMP–AMP synthesis (cGAS) (10). Of these receptors, TLR7, MDA5, and cGAS sense components of Plasmodium either during the liver stage or blood stage of Plasmodium infection (11–13). Upon recognizing pathogen-associated molecular patterns, PRRs trigger downstream signaling activation, inducing the transcription of NF-κB and interferon regulatory factor 3 (IRF3) target genes. Many human cells in the blood, including monocytes, neutrophils, and platelets, take part in fighting malaria parasites (14, 15). Recent studies have shown that the human defense peptide platelet factor 4 (PF4) can directly kill P. falciparum living in infected erythrocytes with assistance from the erythrocyte Duffy antigen receptor (16, 17). Another platelet-secreted chemokine, pro-platelet basic protein (PPBP), a platelet activation marker, also takes part in the process of clearing the parasites by inducing macrophage chemotaxis and mediating neutrophil accumulation (18, 19, 20). Nevertheless, how these chemokines are regulated by upstream signaling pathways, especially innate immune signaling molecules, remains elusive.
The ETS family transcription factors (TFs) are a family of molecules that participate in numerous signaling pathways (21). E74, like ETS transcription factor 4 (ELF4), is one of these TFs and functions in tumorigenesis, DNA damage response, cell cycle regulation, and innate immunity (22–25). In the innate immune system, ELF4 functions as a novel and critical transcription factor of type I IFN, which is of vital importance for host defense against viral infection (25). However, the role of ELF4 in host defense against Plasmodium is unclear.
Here we demonstrate that ELF4 plays a critical role in host defense against Plasmodium yoelii 17XNL. Elf4−/− mice exhibit increased parasitemia compared with WT mice. Expression of Pf4 and Ppbp in Elf4−/− mice is much lower than that of WT mice during infection, without any difference in the number of megakaryocytes or platelets. Our in vitro experiments further reveal that ELF4 can directly bind to the promoter of Pf4 and Ppbp, initiating their transcription and modulating the overall levels of these two chemokines during Plasmodium infection.
Results
Elf4-deficient mice exhibit severe malarial anemia
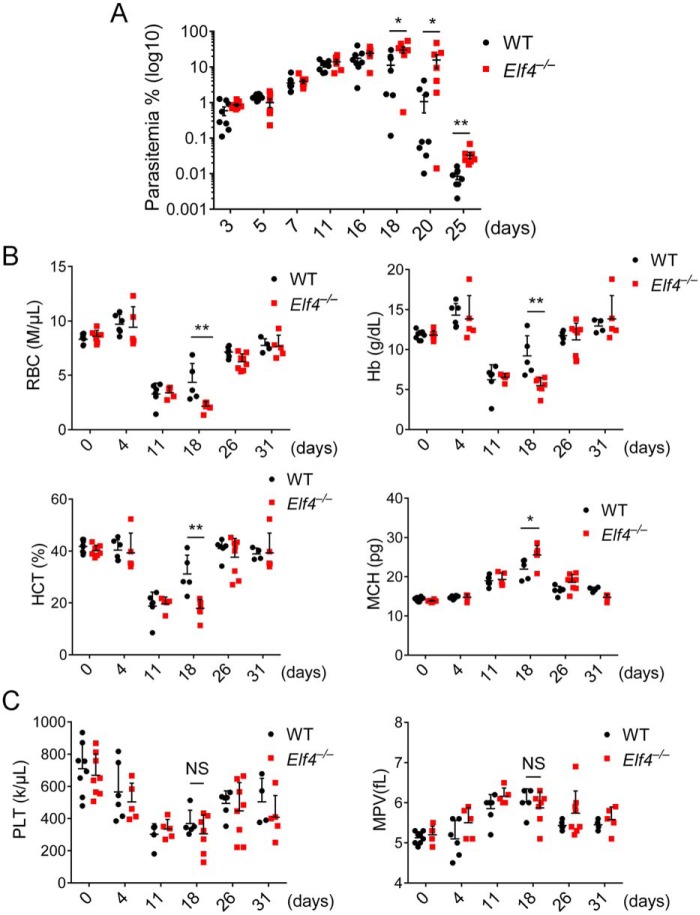
We previously demonstrated that E74, like ELF4, is an important transcription factor of type I IFN and is indispensable for host defense against virus infection (25). However, its function in defense against other pathogens has not been characterized. We sought to determine whether ELF4 plays a role in defense against Plasmodium. Here we used P. yoelii 17XNL, a nonlethal species of P. yoelii, to investigate the function of ELF4 during murine infection with Plasmodium. Elf4+/+ and Elf4−/− mice were injected intraperitoneally with P. yoelii 17XNL–infected red blood cells. Parasitemia peaked on day 18 after infection, when significantly increased parasitemia was observed in Elf4−/− mice and lasted until day 25 (end of parasitemia analysis) (Fig. 1A). Red blood cells (RBCs), hemoglobin (Hb), and the percentage of hematocrit (HCT) during infection indicated the severity of malarial anemia (26), whereas the elevated level of mean corpuscular hemoglobin (MCH) was a marker of hemolytic anemia. Decreased levels of RBCs and Hb together with lower HCT and higher MCH levels were observed in Elf4−/− mice (Fig. 1B). In addition, Elf4−/− mice had elevated neutrophil counts (Fig. S1A) and lower levels of lymphocytes (Fig. S1B). We also observed a higher level of mean corpuscular volume (Fig. S1C). However, there was no difference in platelet counts or mean platelet volume between Elf4+/+ and Elf4−/− mice during infection, indicating a comparable level of thrombocytopoiesis (Fig. 1C). These results showed that Elf4−/− mice exhibited more severe malarial anemia and increased parasitemia compared with WT littermates. These findings establish ELF4 as an indispensable regulator for defense against Plasmodium.
Figure 1.
Elf4-deficient mice suffer from severe malarial anemia. A, parasitemia of Elf4+/+ and Elf4−/− mice infected with GFP–P. yoelii 17XNL (1 × 105 infected red blood cells) on the indicated days after infection was assessed by analyzing the percentage of GFP-positive red blood cells (infected) via flow-cytometry. B, the severity of malarial anemia was assessed by detecting RBCs, Hb, HCT, and MCH levels on the indicated days after infection. C, the level of thrombocytopoiesis was assessed by peripheral platelet (PLT) counts and mean platelet volume (MPV). The data in A are means ± S.E. (n = 8 for Elf4+/+, n = 7 for Elf4−/−). The data in B and C are means ± S.D. All data are from three independent experiments; t test; *, p < 0.05; **p < 0.01; NS, not significant.
Elf4 protects hosts from Plasmodium infection in a type I IFN–independent manner
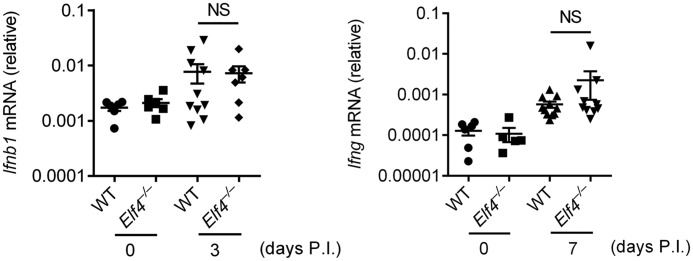
ELF4 is a transcription factor of type I IFN, which is essential for host defense against Plasmodium (27). To investigate the mechanism by which ELF4 mediates anti-Plasmodium defense, we examined whether ELF4 functions through type I IFN. Surprisingly, we did not observe dramatic differences of Ifnb1 expression as well as Ifng after P. yoelii infection (Fig. 2). These data suggest that type I IFN is dispensable for ELF4-mediated host defense against Plasmodium.
Figure 2.
Type I IFN is irrelevant to mediate protection of mice from P. yoelii 17XNL infection. Quantitative PCR analysis of Ifnb1 and Ifng mRNA from blood samples of Elf4+/+ and Elf4−/− mice infected with P. yoelii 17XNL on the indicated days after infection. Gene expression was relative to mouse Hprt. All data are means ± S.D. from three independent experiments; t test; NS, not significant.
The expression of Pf4 and Ppbp is impaired in Elf4-deficient mice
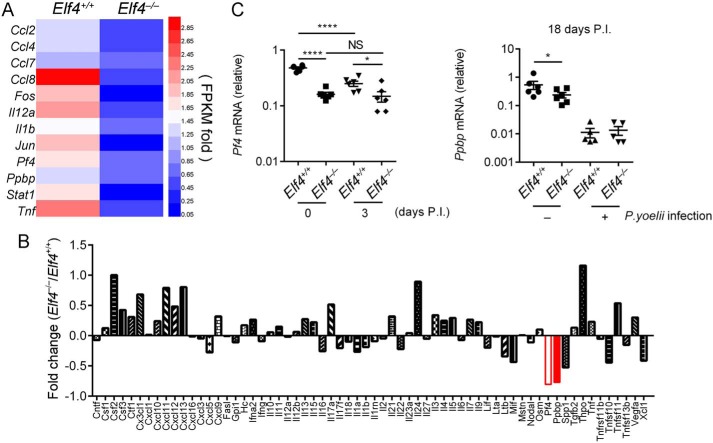
To determine whether anti-Plasmodium effectors are regulated by ELF4, we carried out RNA-Seq to analyze gene expression in the spleen and bone marrow after 2 days of P. yoelii 17XNL infection. Up- and down-regulated genes in Elf4+/+ and Elf4−/− mice were gated via a strategy whereby genes of Elf4−/− samples with more than 1.4-fold FPKM (up-regulated) or less than 0.67-fold FPKM (down-regulated) compared with Elf4+/+ samples were selected to draw the heatmaps (Fig. S2, A and B). Gene Ontology analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis of up- or down-regulated genes suggested major differences in immune response and the cytokine–cytokine receptor interaction process (Fig. S2, C–J). The expression level of Pf4 and Ppbp decreased significantly in the bone marrow of Elf4−/− mice (Fig. 3A). Previous studies have demonstrated that platelets are a critical platform of anti-Plasmodium defense and that PF4 secreted by platelets helps protect against malaria via direct killing of P. falciparum parasites (16). PPBP is known as a platelet activation marker, and the Ppbp gene shares a common megakaryocyte-specific gene locus with Pf4 (28). Consistent with the RNA-Seq results, microarray analysis of whole-blood samples confirmed the down-regulation of Pf4 and Ppbp in Elf4−/− mice (Fig. 3B). To validate the microarray results, qPCR of total blood RNA was used to assess Pf4 and Ppbp mRNA levels (Fig. 3C). Therefore, ELF4 is essential for the production of PF4 and PPBP, which mediate the clearance of Plasmodium.
Figure 3.
Pf4 and Ppbp expression was lower in Elf4−/− mice after P. yoelii 17XNL infection. A, list of the very top enriched genes from RNA-Seq data that were induced more than 1.4-fold in the bone marrow of Elf4+/+ mice compared with Elf4−/− mice. B, microarray analysis of diverse genes expression between Elf4+/+ and Elf4−/− mice in response to infection with P. yoelii 17XNL. C, quantitative PCR analysis of Pf4 and Ppbp mRNA in the blood of Elf4+/+ and Elf4−/− mice on the indicated days after infection with P. yoelii 17XNL. Gene expression in C was relative to mouse Hprt. For C, data are means ± S.D. from three independent experiments; t test; *, p < 0.05; ****, p < 0.0001; NS, not significant.
ELF4 binds to the promoter of Pf4 and Ppbp
The ETS family TFs have highly similar DNA-binding domains, called ETS domains. It is known that ETS family TFs are able to initiate the transcription of megakaryocyte-specific genes (29, 30). Here we reasoned that ELF4 might function by directly binding to promoters of Pf4 and Ppbp. We constructed luciferase plasmids containing the mouse Pf4 or Ppbp promoter region (−1–1000) (Fig. S3A). Remarkably, ELF4, but not other type I IFN signaling molecules, significantly up-regulated the luciferase activity of both mouse Pf4 and Ppbp promoters (Fig. 4A) in a dose-dependent manner (Fig. 4B). We also constructed luciferase plasmids containing the human PF4 or PPBP promoter. ELF4 can also up-regulate human PF4 and PPBP luciferase activity in a dose-dependent manner (Fig. 4C). ELF4 recognizes the target element by its ETS domain, which relies on the DNA-binding activity motif RALR (267–270 amino acids for human ELF4 and 266–269 amino acids for mouse ELF4) (31). Mutation of RALR to AALA (referred to here as ELF4 (AALA)) abolished ELF4 activation on both mouse Pf4 and Ppbp promoters (Fig. 4D) and human PF4 and PPBP promoters (Fig. 4E). These results indicate that ELF4 targets the promoters of Pf4 and Ppbp.
Figure 4.
ELF4 activates the promoter of Pf4 and Ppbp. A and B, luciferase activity in HEK293T cells transfected with mouse Pf4 or Ppbp promoter-driven luciferase reporters together with plasmids encoding FLAG-ELF4, FLAG-MAVS, FLAG-STING, FLAG- IRF3, or FLAG-IRF7 (A) and increasing amounts of plasmids expressing ELF4 (B). C, luciferase activity in HEK293T cells transfected with human PF4 or PPBP promoter-driven luciferase reporters together with increasing amount of plasmids expressing ELF4. D and E, luciferase activity in HEK293T cells transfected with mouse Pf4 or Ppbp promoter-driven (D) or human Pf4 or PPBP promoter-driven (E) luciferase reporters together with plasmids encoding ELF4 or its mutant ELF4(AALA). F, luciferase activity in HEK293T cells transfected with the mouse Pf4 promoter or the different GGAA-mutated, promoter-driven luciferase reporters together with the Elf4 expression plasmid or empty vector. G, luciferase activity in HEK293T cells transfected with different combinations of Pf4 promoter-driven luciferase reporters together with Elf4 expression plasmid or empty vector. H and I, luciferase activity in HEK293T cells transfected with truncation (H) or TTCC- mutation (I) of Pf4 promoter-driven luciferase reporters together with Elf4 expression plasmid or empty vector. J, luciferase activity in HEK293T cells transfected with the mouse Ppbp promoter or the different GGAA-mutated, promoter-driven luciferase reporters together with Elf4 expression plasmid or empty vector. All data are means ± S.D. from three independent experiments; t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We next intended to determine the exact ELF4 binding sites of Pf4 and Ppbp promoters. ELF4 was characterized as a transcription factor mainly binding to the target sequence containing a core purine-rich site, GGAA (31). We identified four GGAA sites in both mouse Pf4 and Ppbp promoters (Fig. S3C). None of the four GGAA sites that were replaced with GATC changed the luciferase activity of the Pf4 promoter (Fig. 4, F and G). Former investigations have found that multiple ETS TFs bind to the GGAA antisense sequence to activate Pf4 expression (30). We thus hypothesize that ELF4 binds to the GGAA antisense sequence (TTCC) of the Pf4 promoter. Because multiple TTCC sites were found in the 1-kb region of the Pf4 promoter, we first built deletion constructs containing 500 bp of the upstream promoter region (−1–500) or a further 500-bp region (−500–1000) to narrow the options of potential sites. Only the −1–500–bp promoter but not the −500–1000–bp promoter luciferase was comparably activated with the full-length promoter by ELF4 (Fig. 4H). We then analyzed the 500-bp Pf4 promoter and found four TTCC sites. Mutation of the −5 TTCC site but not the other three TTCC sites resulted in disability of luciferase activation by ELF4 (Fig. 4I), suggesting that the −5 GGAA antisense TTCC site was the ELF4-binding site of the Pf4 promoter. Compared with the Pf4 promoter, activation of the Ppbp promoter luciferase was impaired when the first GGAA (−702) was replaced with GATC (Fig. 4J). We also intended to find the ELF4-binding sites on the human PF4 and PPBP promoters. We constructed human PF4 and PPBP promoter luciferase plasmids. Following the same strategy we used to find the binding sites on mouse promoters, we identified the −305 GGAA of the PF4 promoter (Fig. S4B) and −57 TTCC of the PPBP promoter (Fig. S4C) as ELF4-binding sites.
Although a predicted promoter of genes usually locates at the upstream region of the transcriptional start site (TSS), there remains the possibility that some noncoding exons localized upstream of already known TSSs could also be transcribed in specific tissues or cells (e.g. in megakaryocytes). Given that the −1.1-kb region upstream of rat PF4 is the promoter of rat PF4 in megakaryocytes (32), we wanted to make sure that the 1-kb region we used to build luciferase constructs contained the true promoters of mouse Pf4 and Ppbp in megakaryocytes. We designed primers targeting the −200, −400, or −1000 region upstream of the TSS of Pf4 or Ppbp. We also designed the +1–93 primer or +1–249 primer of Pf4 or Ppbp. Quantitative PCR (qPCR) (Fig. S3, D and E) or RT-PCR (Fig. S3, F and G) using cDNA or genomic DNA of megakaryocytes isolated from WT C57BL/6 mice demonstrated that the upstream region of the TSS was not included in the heterogeneous nuclear RNA of Pf4 or Ppbp. Taken together, our results demonstrate that ELF4 binds to Pf4 and Ppbp promoters.
ELF4 promotes the expression of Pf4 and Ppbp
We already discovered that ELF4 binds to the promoter of Pf4 and Ppbp. Next we wanted to determine the effects of ELF4 on the expression of Pf4 and Ppbp. To evaluate the effects of ELF4 expression on PF4 and PPBP transcriptional activation, we used megakaryocytic HEL (human erythroleukemia) cells. We generated ELF4-KO HEL cells using the CRISPR-Cas9 system. Using a lentivirus packaging system, we also generated ELF4- or ELF4(AALA)-expressing HEL cells. HEL cells stably expressing ELF4 expressed higher levels of PF4 and PPBP (Fig. 5A), whereas endogenous expression of PF4 and PPBP was much lower in ELF4-KO HEL cells (Fig. 5B). To prove that ELF4 binds directly to the promoter of PF4 and PPBP, we performed a ChIP assay in HEL cells with or without ELF4 or ELF4(AALA) expression. ELF4, but not the ELF4(AALA) mutant, directly bound the PF4 and PPBP promoter (Fig. 5C). We also used another megakaryocytic cell line, Dami cells, to further verify our hypothesis. Overexpressed ELF4, but not the ELF4(AALA) mutant, up-regulated PF4 and PPBP expression (Fig. S4A) and bound to the promoter region of both PF4 and PPBP (Fig. S4B) in Dami cells. Taken together, these results suggest that ELF4 binds directly to PF4 and PPBP promoters and promotes the expression of PF4 and PPBP.
Figure 5.
ELF4 mediates the transcriptional activation of Pf4 and Ppbp. A, expression of PF4 and PPBP in WT HEL cells or HEL cells stably transfected with ELF4 was assessed by qPCR analysis. VEC, vector. cpVEC, lentivirus packaging lentiCRISPRv2-vector infected. B, expression of PF4 and PPBP in WT or ELF4-KO HEL cells was assessed by qPCR analysis. C, the binding ability of ELF4 on PF4 and PPBP promoters was assessed by ChIP-qPCR analysis in WT HEL cells or HEL cells stably transfected with ELF4 or ELF4(AALA). For A and B, gene expression was relative to human GAPDH. All data are means ± S.D. from three independent experiments; t test; *, p < 0.05; **, p < 0.01.
ELF4 is essential for controlling Pf4 and Ppbp expression during P. yoelii infection
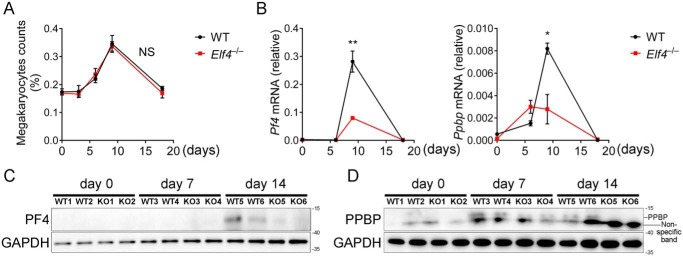
Bone marrow megakaryocytes are the main source of PF4 and PPBP in peripheral platelets. Although malarial anemia caused by P. yoelii infection could result in increased numbers of megakaryocytes and decreased numbers of thromocytogenic megakaryocytes, we observed no obvious differences in the quantity of bone marrow megakaryocytes (Fig. 6A) or peripheral platelets (Fig. 1C) in Elf4+/+ and Elf4−/− mice. Because platelet-secreted PF4 and PPBP mediate clearance of Plasmodium, and ELF4 activates Pf4 and Ppbp, we hypothesize that ELF4 is essential for controlling the expression of Pf4 and Ppbp in megakaryocytes. To determine the ongoing ability of ELF4 to activate Pf4 and Ppbp during Plasmodium infection, we analyzed the expression of Pf4 and Ppbp in bone marrow megakaryocytes and peripheral platelets, respectively. We isolated bone marrow megakaryocytes and peripheral platelets to assess the expression of Pf4 and Ppbp during P. yoelii infection. Gene expression of Pf4 first increased and then reduced and was not significantly up-regulated in megakaryocytes until day 9 after infection, when Pf4 was expressed at a lower level in Elf4−/− mice. During Plasmodium infection, the expression of Ppbp exhibited a similar tendency with Pf4 (Fig. 6B). Immunoblot analysis indicated that the protein levels of PF4 (Fig. 6C) and PPBP (Fig. 6D) decreased in peripheral platelets of Elf4−/− mice. These data indicated that the lower level of Pf4 and Ppbp expression in Elf4−/− mice was not due to the decreased number of megakaryocytes or platelets. Taken together, these results suggest that ELF4 plays an important role in modulating the expression of Pf4 and Ppbp during Plasmodium infection.
Figure 6.
ELF4 modulates the expression of Pf4 and Ppbp during P. yoelii infection. A, quantification of bone marrow megakaryocytes was assessed by calculating the percentage of megakaryocytes in total bone marrow cells on the indicated days after infection. B, expression of Pf4 and Ppbp in isolated megakaryocytes from Elf4+/+ or Elf4−/− mice on day 0, 6, 9, or 18 after infection. C, protein level of Pf4 in peripheral platelets on day 0, 7, or 14 after P. yoelii infection from individual Elf4+/+ or Elf4−/− mice. D, protein level of Ppbp in peripheral platelets on day 0, 7, or 14 after P. yoelii infection from the same individual Elf4+/+ or Elf4−/− mice as in C. For B, gene expression was relative to Actb. For C and D, GAPDH was used as a control. The data in A and B are means ± S.D. (n = 3). All data are from two independent experiments; t test; *, p < 0.05; **, p < 0.01; NS, not significant.
Discussion
Diverse studies have examined how the immune system helps to defend against malaria (33). Host defense against malaria mainly includes innate immunity, adaptive immunity, and other mechanisms to kill parasites. Recent studies have found that certain innate immune molecules, such as MAVS (mitochondrial antiviral signaling protein), STING (stimulator of interferon genes), and cGAS, suppressed host defense against Plasmodium, resulting in unexpectedly severe malaria. Different from their role in antiviral immunity, Mb21d1 (cGAS)–, Sting-, Mda5-, Mavs-, or transcription factor Irf3–deficient mice produced high amounts of type I interferon in the serum and were resistant to lethal P. yoelii YM infection (13). Although the innate immune pathway involved in anti-Plasmodium defense has been well studied, there is no explicit innate signaling molecule that can function against Plasmodium infection independently of type I interferon. ELF4 is an ETS domain transcription factor essential for production of type I interferon and antiviral immunity during virus infection. Here we demonstrated that ELF4 mediated anti-Plasmodium immunity in a type I interferon–independent fashion. ELF4 binds directly to the promoter and facilitates the transcription of Pf4 and Ppbp, which help to kill Plasmodium directly.
Accumulating evidence demonstrates that ELF4 regulates blood cell proliferation (21). When we analyzed cells in the blood, we found elevated numbers of reticulocytes in Elf4−/− mice (data not shown). Because P. yoelii 17XNL prefers to invade reticulocytes but not erythrocytes (34), Elf4 deficiency provides an ideal environment for parasite infection. Because of the preference of Plasmodium to invade reticulocytes, more uninfected erythrocytes presenting phosphatidylserine will be opsonized by macrophages (35), resulting in aggravating anemia. Elevated reticulocytes can also result in a higher relative HCT percentage, which may explain the unexpectedly higher level of mean corpuscular volume.
Megakaryocyte-secreted PF4 can regulate hematopoietic stem cell quiescence (36). Previous studies have shown that PF4 promotes monocyte activation and subsequent cytokine production via increasing Klf4 expression in monocytes, resulting in severe cerebral inflammation in experimental cerebral malaria (17), which may be avoided because of ELF4 deficiency. These studies demonstrate that ELF4 may function via PF4 to regulate various kinds of blood cells. Ppbp shares a common proximal location in the genome with Pf4. These two cytokines are expressed during a similar period during megakaryocyte development (28). PPBP functions through two steps of N-terminal cleavage, forming neutrophil-activating peptide 2 (NAP-2), an antibacterial protein functioning in the innate immune system (37). However, we observed equal levels of thrombocytopoiesis in Elf4+/+ and Elf4−/− mice during P. yoelii infection in our study, indicating the need for a detailed examination of changes in different cell types to uncover the more complicated mechanism in which ELF4 plays a role in the P. yoelii infection model. In summary, although ELF4 directly promotes transcription of Pf4 and Ppbp, how ELF4 fine-tunes anti-Plasmodium immunity remains to be investigated further.
We found previously that ELF4 is involved in innate immune signaling. ELF4 is recruited by STING and activated by tumor necrosis factor receptor-associated factor (TRAF) family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1). A recent study showed that STING and TBK1 suppress anti-Plasmodium immunity (13). It is interesting to explore whether these two molecules may regulate the expression of PF4 and PPBP. In conclusion, our study showed that the innate immune signaling molecule ELF4 functions in defending against P. yoelii infection via activating transcription Pf4 and Ppbp.
Experimental procedures
Ethics statement
This study was carried out in accordance with the recommendations of National Regulations for the Administration of Affairs Concerning Experimental Animals. The protocol was approved by the Peking University Animal Care and Use Committee. The project license number is LA2016239.
Malaria parasites and mice
The rodent parasite P. yoelii 17XNL strain used in this study was kindly provided by Dr. J. Yuan (School of Life Science, Xiamen University). GFP-expressing P. yoelii 17XNL strain used in this study was initially obtained from the Malaria Research and Reference Reagent Resource Center. P. yoelii parasites were thawed from frozen stocks and maintained alive by continuous intraperitoneal passaging in mice every 6 days. Elf4−/− mice on a C57BL/6J background have been described previously (24). All mice were housed under specific pathogen–free conditions, and all mice were analyzed at 6–8 weeks of age (unless otherwise specified).
Infection of mice with P. yoelii parasites
An inoculum containing appropriate numbers of infected red blood cells (1 × 105) suspended in 100 μl of PBS (pH 7.4) from donor mice was injected i.p. into experimental C57BL/6J knockout mice or littermates. Mouse blood was collected on the indicated day to monitor parasitemia. Parasitemia was analyzed by flow cytometry. For flow cytometry analysis using FACSVerse (BD Biosciences), a drop of blood in PBS was used to measure blood parasitemia. GFP-expressing parasites were detected in the green fluorescent channel FL1. The gated amount related to all detected live cells corresponded to blood parasitemia in percent.
Blood indices
Blood was obtained on the indicated day after infection by tail snip. 20 μl of blood was collected in a heparin-coated tube and then analyzed with an HEM7AVET 950 analyzer (Drew Scientific Inc.).
Platelet isolation
Whole blood of mice was collected from the retro-orbital sinus into EDTA-containing microtubes. Platelet-rich plasma was obtained by centrifugation of whole blood at 110 × g for 10 min. Platelets were isolated by further centrifugation at 800 × g for 10 min and lysed using radioimmune precipitation assay lysis buffer to perform the subsequent immunoblot assay.
Bone marrow collection and megakaryocyte purification
Bone marrow was collected from the tibiae and femora of mice on the indicated days after P. yoelii infection. Megakaryocytes were purified using a mouse megakaryocyte isolation kit (MEG2014M, TBD Sciences). Briefly, bone marrow was resuspended and passed through a 70-μm nylon cell strainer, followed by Percoll gradient (63/30%) centrifugation. Megakaryocytes were harvested from the top interphase. Collected megakaryocytes were washed with PBS twice and used to purify total RNA by RNeasy Mini Kit (Qiagen).
Determination of megakaryocyte number
Quantification of megakaryocytes was performed via bone marrow megakaryocyte isolation and counting. Briefly, the bone marrow was flushed out using PBS, resuspended, and passed through a 70-μm nylon cell strainer, followed by total bone marrow cell counting. After the megakaryocytes were isolated, the collected cells were counted, and the percentage of megakaryocytes in total bone marrow cells was calculated.
Cell culture
HEK293T cells were maintained in DMEM (Gibco) with 10% FBS and 1% glutamine (Gibco). HEL and Dami cells were cultured in RPMI 1640 medium (Gibco) with 10% FBS and 1% glutamine (Gibco). All cells were incubated at 37 °C with 5% CO2. Cells tested negative for mycoplasma contamination.
Generation of stable cell lines
ELF4−/− HEL cells and Dami cells or HEL cells stably expressing FLAG-ELF4 or ELF4 (AALR) were generated as follows. For ELF4-KO cells, single guide RNA oligos (targeting sequence, 5′-GAGTTGGACGACGTTCACAA-3′) were cloned into lentiCRISPRv2 following the lentiCRISPRv2 oligo cloning protocol. For ELF4 overexpression, FLAG-ELF4 (or ELF4(AALA)) was amplified by PCR and cloned into the pCDH-CMV-MCS-EF1-puro vector. Lentiviral vectors (lentiCRISPRv2 or pCDH-CMV-MCS-EF1-puro) were co-transfected with the packaging plasmids pVSVg and psPAX2 into HEK293T cells for 48 h. Culture supernatants containing viral particles were collected and filtered through a 0.45-μm nitrocellulose filter (Millipore). Cells were infected in the presence of 4 μg/ml Polybrene and selected with 10 μg/ml puromycin. All stable cell lines were used at early passages.
Reporter assay
HEK293T cells (2 × 105) were plated in 24-well plates and transfected using PEI (Polysciences) with plasmids encoding Pf4 or Ppbp luciferase reporter (firefly luciferase, 100 ng) and pRL-TK (Renilla luciferase plasmid, 10 ng) together with 100 ng of plasmid encoding FLAG-ELF4, FLAG-ELF4(AALA), FLAG-MAVS, FLAG-STING, FLAG-IRF3, or FLAG-IRF7. Deletion or GGAA (TTCC) mutation of the mouse Pf4 and Ppbp (or human PF4 and PPBP) promoter region containing luciferase plasmids was carried out using standard molecular biology methods. The empty p3×FLAG-CMV-7.1 vector was used to maintain equal amounts of DNA among wells. After transfection for 24 h, cells were lysed, and luciferase activity was measured with the Dual-Luciferase Assay System (Promega) according to the manufacturer's instructions. Reporter gene activity was determined by normalization of firefly luciferase activity to Renilla luciferase activity.
RT-PCR and quantitative real-time PCR
Total RNA was extracted from tissue or cells using the RNeasy Mini kit (Qiagen), and the cDNA was generated using reverse transcriptase III (Invitrogen). For RT-PCR, 1 μl of RT reaction was used for each PCR, and the number of cycles was optimized to avoid saturation (32 for all the primer pairs). 3 μl of reaction product was loaded on agarose gels. Real-time PCR was carried out using the 7500 Fast Real-Time PCR System (Applied Biosystems) using SYBR GreenER qPCR Super Mix Universal (Invitrogen) and specific primers. The relative RNA expression level was normalized to Hprt, Actb (for mouse), or GAPDH (for human) according to the 2−ΔCt calculation method. Primer pairs used to detect target gene transcripts are listed in Table S1.
Transcriptomics analysis
The total RNA of blood samples was purified using the RNeasy Mini Kit (Qiagen). RNA quantity was evaluated spectrophotometrically, and RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies). All samples showed an RNA integrity number of more than 8. RNA-Seq libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) and sequenced on an Illumina Hiseq platform. Sequencing was performed at Beijing Novogene Bioinformatics Technology Co. Ltd. and Nanjing Vazyme Biotech Co. Ltd. The filtered reads were mapped to the mouse genome reference sequence (GRCm38/mm10 Ensemble release 83) using HISAT2. Gene expression was quantified as fragments per kilobase of transcript per million fragments mapped (FPKM) algorithm. Genes with an FPKM value of more than 1.4-fold or less than 0.67-fold were assigned as differentially expressed. Network visualization of Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway mapping was performed with DAVID 6.7. Heatmaps were generated using HEML 1.0 (Heatmap Illustrator, version 1.0 (38)). See Tables S2 and S3 for more details regarding gene list and analysis results.
Microarray analysis
Peripheral blood samples were collected in heparin-coated tubes. RNA was extracted using the RNeasy Mini Kit (Qiagen), and DNase treatment was performed with DNase I (Sigma). The complementary DNA was subsequently synthesized. Labeled cDNA (using Cyanine-3) was synthesized, purified, quantified, and prepared for hybridization following the Agilent protocol. The microarray experiment was performed on a one-color 4 × 180,000 oligonucleotide array (Agilent Technologies, G4839A) according to the manufacturer's protocol. The microarray slides were preprocessed (signal background corrections) and scanned using Agilent's High-Resolution C scanner. Following normalization, the probe intensity of all probes in a probe set was summarized to a single value. The bulk of the analyses were done on the RMA (robust multi-array average)-normalized data in log2 scale.
Immunoblotting
Whole-cell extracts were prepared using radioimmune precipitation assay lysis buffer (50 mm Tris (pH 7.4), 150 mm NaCl, 1% NP-40, 0.1% SDS, and 0.5% deoxycholate supplemented with a protease inhibitor tablet (Roche)) and centrifuged at 10,000 × g for 10 min at 4 °C. The protein concentration was measured using Pierce BCA protein assay kit (Thermo Fisher Scientific). Proteins were resolved on 15% SDS-PAGE and transferred onto nitrocellulose membranes (Millipore). Blocked membranes were incubated with primary antibodies (rabbit anti-PF4 (Proteintech), rabbit anti-PPBP (Sigma-Aldrich), and mouse anti-GAPDH (Proteintech)). Secondary antibodies conjugated to HRP and Western chemiluminescent HRP substrate (Millipore) were used for detection.
ChIP assay
HEL or Dami cells (1.5 × 107) in 10-cm dishes were cross-linked with 1% formaldehyde for 10 min at room temperature. The reaction was quenched by adding glycine solution (final concentration, 0.1 m) and then incubating for 5 min. Cells were then washed with ice-cold PBS with PMSF and resuspended in lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris-HCl (pH 8.1) supplemented with a protease inhibitor tablet (Roche)). Chromatin was sheared by sonication on ice to generate 200- to 1000-bp chromatin fragments, and the insoluble fraction was removed by centrifugation. The lysate was precleared with protein A/G beads (Invitrogen) and then incubated with anti-FLAG affinity gel (B23102, Bimake Biotech) overnight at 4 °C. Immunocomplexes were collected and washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 7.9), and 150 mm NaCl), high salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 7.9), and 500 mm NaCl), LiCl wash buffer (0.25 m LiCl, 0.5 mm sodium deoxycholate, 1 mm EDTA, 0.5% NP40, and 10 mm Tris-HCl (pH 8.0)), and TE buffer (10 mm Tris-HCl (pH 7.9) and 1 mm EDTA). Precipitated chromatin fragments were eluted using elution buffer (1% SDS and 0.1 m NaHCO3). After adding NaCl (final concentration, 0.2 m), samples were reverse-cross-linked at 65 °C overnight. After proteinase K (GE201, TransGen Biotech) digestion, DNA was extracted using phenol/chloroform and purified for quantitative real-time PCR analyses using specific primers (see Table S1 for primer sequences).
Statistical analysis
All analyses were performed using GraphPad Prism version 6.0 (GraphPad Software, La Jolla, CA). Data are presented as means ± S.D. unless stated otherwise. Statistical significance of differences between two groups was assessed by unpaired Student's t tests, and p < 0.05 was considered significant.
Author contributions
D. W., Z. Z., E. F., and F. Y. conceptualization; D. W., Z. Z., S. Craft, and F. Y. data curation; D. W., Z. Z., S. Cui, Y. Z., and S. Craft software; D. W., Z. Z., and Y. Z. formal analysis; D. W., Z. Z., S. Cui, and F. Y. validation; D. W., Z. Z., S. Cui, Y. Z., S. Craft, E. F., and F. Y. investigation; D. W., Z. Z., S. Cui, Y. Z., S. Craft, and F. Y. methodology; D. W. and Z. Z. writing-original draft; Z. Z., S. Cui, E. F., and F. Y. resources; Z. Z., E. F., and F. Y. supervision; Z. Z., E. F., and F. Y. writing-review and editing; S. Craft, E. F., and F. Y. project administration; E. F. and F. Y. funding acquisition.
Supplementary Material
Acknowledgment
We thank Dr. J. Yuan (School of Life Science, Xiamen University) for providing P. yoelii 17XNL.
This work was supported by National Natural Science Foundation of China Grant 31570891 and 31872736 National Key Research and Development Program of China Grant 2016YFA0500302. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1–S3.
- PRR
- pattern recognition receptor
- cGAS
- cGMP–AMP synthesis
- PPBP
- pro-platelet basic protein
- ETS
- erythroblast transformation specific
- TF
- transcription factor
- RBC
- red blood cell
- Hb
- hemoglobin
- HCT
- hematocrit
- MCH
- mean corpuscular hemoglobin
- FPKM
- fragments per kilobase of transcript per million fragments mapped
- qPCR
- quantitative PCR
- TSS
- transcription start site
- cDNA
- complementary DNA
- HEL cells
- human erythroleukemia cells
- MAVS
- mitochondrial antiviral signaling protein
- STING
- stimulator of interferon genes.
References
- 1. Murray C. J. L., Rosenfeld L. C., Lim S. S., Andrews K. G., Foreman K. J., Haring D., Fullman N., Naghavi M., Lozano R., and Lopez A. D. (2012) Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379, 413–431 10.1016/S0140-6736(12)60034-8 [DOI] [PubMed] [Google Scholar]
- 2. Cowman A. F., Healer J., Marapana D., and Marsh K. (2016) Malaria: biology and disease. Cell 167, 610–624 10.1016/j.cell.2016.07.055 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization (2015) Guidelines for the treatment of malaria, 3rd Ed., http://www.who.int/malaria/publications/atoz/9789241549127/en/ [PubMed]
- 4. Noedl H., Se Y., Schaecher K., Smith B. L., Socheat D., Fukuda M. M., and Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium (2008) Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med 359, 2619–2620 10.1056/NEJMc0805011 [DOI] [PubMed] [Google Scholar]
- 5. Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., Lwin K. M., Ariey F., Hanpithakpong W., Lee S. J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., et al. (2009) Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361, 455–467 10.1056/NEJMoa0808859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 7. Barrat F. J., Elkon K. B., and Fitzgerald K. A. (2016) Importance of nucleic acid recognition in inflammation and autoimmunity. Annu. Rev. Med. 67, 323–336 10.1146/annurev-med-052814-023338 [DOI] [PubMed] [Google Scholar]
- 8. Kawai T., and Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 9. Rehwinkel J., and Reis e Sousa C. (2010) RIGorous detection: exposing virus through RNA sensing. Science 327, 284–286 10.1126/science.1185068 [DOI] [PubMed] [Google Scholar]
- 10. Sun L., Wu J., Du F., Chen X., and Chen Z. J. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gazzinelli R. T., Kalantari P., Fitzgerald K. A., and Golenbock D. T. (2014) Innate sensing of malaria parasites. Nat. Rev. Immunol. 14, 744–757 10.1038/nri3742 [DOI] [PubMed] [Google Scholar]
- 12. Liehl P., Zuzarte-Luís V., Chan J., Zillinger T., Baptista F., Carapau D., Konert M., Hanson K. K., Carret C., Lassnig C., Müller M., Kalinke U., Saeed M., Chora A. F., Golenbock D. T., et al. (2014) Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20, 47–53 10.1038/nm.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu X., Cai B., Wang M., Tan P., Ding X., Wu J., Li J., Li Q., Liu P., Xing C., Wang H. Y., Su X. Z., and Wang R. F. (2016) Cross-regulation of two type I interferon signaling pathways in plasmacytoid dendritic cells controls anti-malaria immunity and host mortality. Immunity 45, 1093–1107 10.1016/j.immuni.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Souza M. C., Padua T. A., Henriques M. G. (2015) Endothelial-leukocyte interaction in severe malaria: beyond the brain. Mediators Inflamm. 2015, 168937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Palomo J., Quesniaux V. F. J., Togbe D., Reverchon F., and Ryffel B. (2018) Unravelling the roles of innate lymphoid cells in cerebral malaria pathogenesis. Parasite Immunol. 40 10.1016/j.chom.2012.10.017 [DOI] [PubMed] [Google Scholar]
- 16. Love M. S., Millholland M. G., Mishra S., Kulkarni S., Freeman K. B., Pan W., Kavash R. W., Costanzo M. J., Jo H., Daly T. M., Williams D. R., Kowalska M. A., Bergman L. W., Poncz M., DeGrado W. F., et al. (2012) Platelet factor 4 activity against P. falciparum and its translation to nonpeptidic mimics as antimalarials. Cell Host Microbe 12, 815–823 10.1111/pim.12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srivastava K., Cockburn I. A., Swaim A., Thompson L. E., Tripathi A., Fletcher C. A., Shirk E. M., Sun H., Kowalska M. A., Fox-Talbot K., Sullivan D., Zavala F., and Morrell C. N. (2008) Platelet factor 4 mediates inflammation in experimental cerebral malaria. Cell Host Microbe 4, 179–187 10.1016/j.chom.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brouwers J., Noviyanti R., Fijnheer R., de Groot P. G., Trianty L., Mudaliana S., Roest M., Syafruddin D., van der Ven A., and de Mast Q. (2014) Platelet activation determines angiopoietin-1 and VEGF levels in malaria: implications for their use as biomarkers. PLoS ONE 8, e64850 10.1371/journal.pone.0064850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Unver N., Esendagli G., Yilmaz G., and Guc D. (2015) CXCL7-induced macrophage infiltration in lung tumor is independent of CXCR2 expression CXCL7-induced macrophage chemotaxis in LLC tumors. Cytokine 75, 330–337 10.1016/j.cyto.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 20. Brown A. J., Sepuru K. M., Sawant K.V., and Rajarathnam K. (2017) Platelet-derived chemokine CXCL7 dimer preferentially exists in the glycosaminoglycan-bound form: implications for neutrophil-platelet crosstalk. Front. Immunol. 8, 1248 10.3389/fimmu.2017.01248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suico M. A., Shuto T., and Kai H. (2017) Roles and regulations of the ETS transcription factor ELF4/MEF. J. Mol. Cell. Biol. 9, 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lacorazza H. D., Yamada T., Liu Y., Miyata Y., Sivina M., Nunes J., and Nimer S. D. (2006) The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9, 175–187 10.1016/j.ccr.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 23. Yamada T., Park C. S., Mamonkin M., and Lacorazza H. D. (2009) Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat. Immunol. 10, 618–626 10.1038/ni.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lacorazza H. D., Miyazaki Y., Di Cristofano A., Deblasio A., Hedvat C., Zhang J., Cordon-Cardo C., Mao S., Pandolfi P. P., Nimer S. D. (2002) The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17, 437–449 10.1016/S1074-7613(02)00422-3 [DOI] [PubMed] [Google Scholar]
- 25. You F., Wang P., Yang L., Yang G., Zhao Y. O., Qian F., Walker W., Sutton R., Montgomery R., Lin R., Wasaki A., and Fikrig E. (2013) ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol. 14, 1237 10.1038/ni.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu L., Zheng X., Berzins K., and Chaudhuri A. (2013) Cytokine dysregulation associated with malarial anemia in Plasmodium yoelii infected mice. Am. J. Transl. Res. 5, 235–245 [PMC free article] [PubMed] [Google Scholar]
- 27. Wu J., Cai B., Sun W., Huang R., Liu X., Lin M., Pattaradilokrat S., Martin S., Qi Y., Nair S. C., Bolland S., Cohen J. I., Austin C. P., Long C. A., Myers T. G., et al. (2015) Genome-wide analysis of host-Plasmodium yoelii interactions reveals regulators of the type I interferon response. Cell Rep. 12, 661–672 10.1016/j.celrep.2015.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang C., Thornton M. A., Kowalska M. A., Sachis B. S., Feldman M., Poncz M., McKenzie S. E., and Reilly M. P. (2001) Localization of distal regulatory domains in the megakaryocyte-specific platelet basic protein/platelet factor 4 gene locus. Blood 98, 610–617 10.1182/blood.V98.3.610 [DOI] [PubMed] [Google Scholar]
- 29. Zhang C., Gadue P., Scott E., Atchison M., and Poncz M. (1997) Activation of the megakaryocyte-specific gene platelet basic protein (PBP) by the Ets family factor PU.1. J. Biol. Chem. 272, 26236–26246 10.1074/jbc.272.42.26236 [DOI] [PubMed] [Google Scholar]
- 30. Okada Y., Nobori H., Shimizu M., Watanabe M., Yonekura M., Nakai T., Kamikawa Y., Wakimura A., Funahashi N., Naruse H., Watanabe A., Yamasaki D., Fukada S., Yasui K., Matsumoto K., et al. (2011) Multiple ETS family proteins regulate PF4 gene expression by binding to the same ETS binding site. PLoS ONE 6, e24837 10.1371/journal.pone.0024837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A. R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H. G., et al. (2010) Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 10.1038/emboj.2010.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ravid K., Beeler D. L., Rabin M. S., Ruley H. E., and Rosenberg R. D. (1991) Selective targeting of gene products with the megakaryocyte platelet factor 4 promoter. Proc. Natl. Acad. Sci. U.S.A. 88, 1521–1525 10.1073/pnas.88.4.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Head M. G., Goss S., Gelister Y., Alegana V., Brown R. J., Clarke S. C., Fitchett J. R. A., Atun R., Scott J. A. G., Newell M. L., et al. (2017) Global funding trends for malaria research in sub-Saharan Africa: a systematic analysis. Lancet Glob. Health 5, e772–e781 10.1016/S2214-109X(17)30245-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vigário A. M., Belnoue E., Cumano A., Marussig M., Miltgen F., Landau I., Mazier D., Gresser I., and Rénia L. (2001) Inhibition of Plasmodium yoelii blood-stage malaria by interferon α through the inhibition of the production of its target cell, the reticulocyte. Blood 97, 3966–3971 10.1182/blood.V97.12.3966 [DOI] [PubMed] [Google Scholar]
- 35. Fernandez-Arias C., Rivera-Correa J., Gallego-Delgado J., Rudlaff R., Fernandez C., Roussel C., Götz A., Gonzalez S., Mohanty A., Mohanty S., Wassmer S., Buffet P., Ndour P. A., and Rodriguez A. (2016) Anti-self phosphatidylserine antibodies recognize uninfected erythrocytes promoting malarial anemia. Cell Host Microbe 19, 194–203 10.1016/j.chom.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bruns I., Lucas D., Pinho S., Ahmed J., Lambert M. P., Kunisaki Y., Scheiermann C., Schiff L., Poncz M., Bergman A., and Frenette P. S. (2014) Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 20, 1315–1320 10.1038/nm.3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krijgsveld J., Zaat S. A., Meeldijk J., van Veelen P. A., Fang G., Poolman B., Brandt E., Ehlert J. E., Kuijpers A. J., Engbers G. H., Feijen J., and Dankert J. (2000) Thrombocidins, microbicidal proteins from human blood platelets, are C-terminal deletion products of CXC chemokines. J. Biol. Chem. 275, 20374–20381 10.1074/jbc.275.27.20374 [DOI] [PubMed] [Google Scholar]
- 38. Deng W., Wang Y., Liu Z., Cheng H., and Xue Y. (2014) HemI: a toolkit for illustrating heatmaps. PLoS ONE 9, e111988 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.