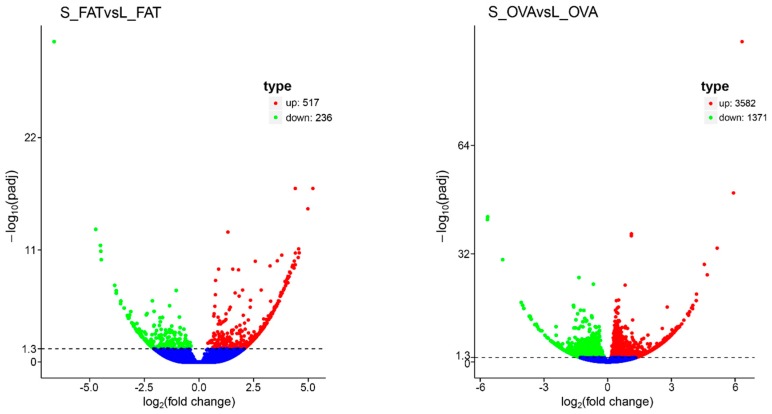
Figure 1.
DEGs between long and short photoperiods treatments in L. migratoria fat body (left) and ovary (right) samples. DEGSeq (2010) R package (1.10.1) was used to carry out the differential expression analysis in digital gene expression and determining the expression via model based negative binomial distribution. Resulting p values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value of <0.05 explained by DEGSeq were assigned as differentially expressed. The x-axis represents the change of gene expression in different groups; whereas the y-axis represents the statistical significance of gene expression change. −log10(padj) means −log10 (adjusted p-value). The smaller the adjusted p-value in −log10(padj), the greater the difference will be (significant). Blue dots in the figure represent the genes with no significant difference; red dots represent the up-regulated genes with significant difference, whereas green dots represent the down-regulated genes with significant difference.