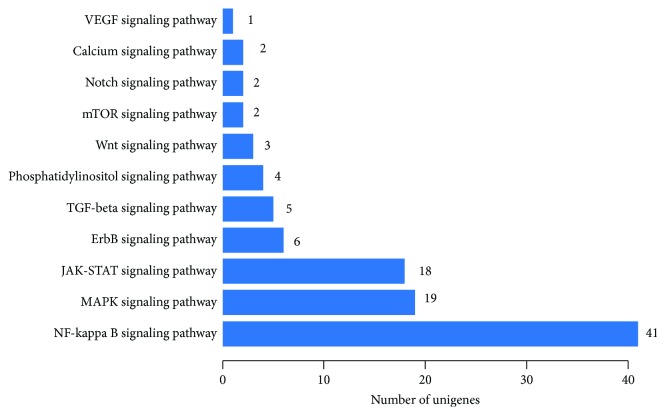
Figure 3.
LPS mainly activated the NF-κB, MAPK, and JAK-ATAT signaling pathways of bMEC. The number of differentially expressed genes relates to cell signaling transduction in LPS-stimulated bMEC. The total RNA was prepared after stimulation with LPS or without any processing for 12 h; then, the RNA-seq experiment and transcriptome assembly analysis were performed by BGI Tech. With significant pathway enrichment, we can ascertain the main biochemical pathways and signal transduction pathways which differentially expressed genes take part in. All the genes were used for the KEGG ontology (KO) enrichment analyses. For KO enrichment analysis, a Q value ≤ 0.05 was used as the threshold to determine significant enrichment of the gene sets.