Figure 2.
EGFR Controls a Variety of Genes Required for Epidermal Differentiation and DNA Integrity
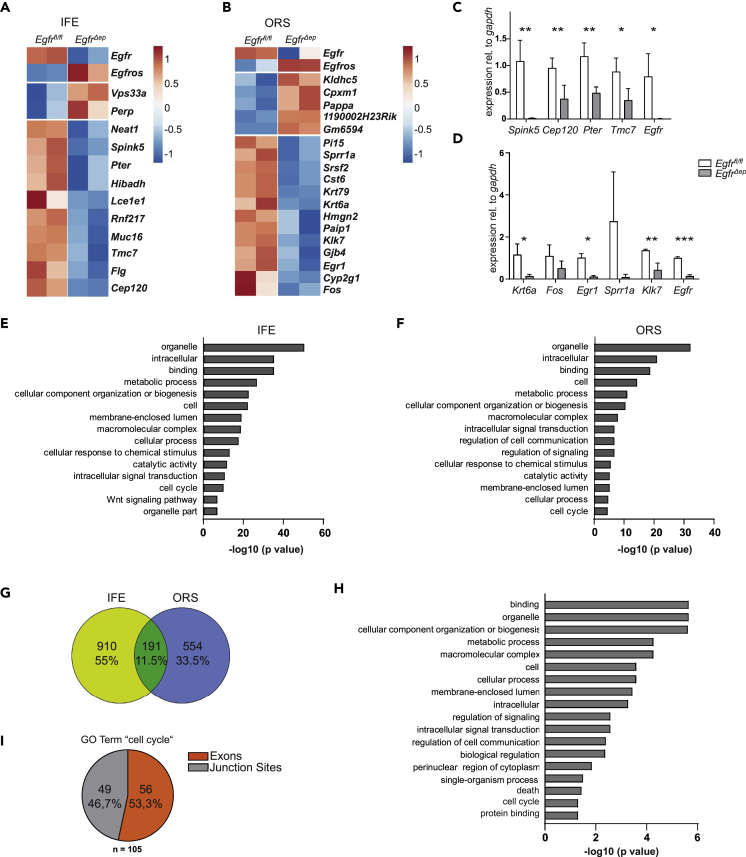
(A) Heatmap of significantly deregulated genes in P2 fluorescence-activated cell sorted IFE from control and EgfrΔep mice. Each column represents one mouse.
(B) Heatmap of significantly deregulated genes in P2 fluorescence-activated cell sorted ORS from control and EgfrΔep mice. Each column represents one mouse.
(C) qRT-PCR from randomly chosen target genes identified by RNA-seq from P2 IFE. Bars show data from n = 3–5 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05, **p < 0.01 as determined by Student's t test.
(D) qRT-PCR from randomly selected target genes identified by RNA-seq from P2 ORS. Bars show data from n = 3–5 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05, **p < 0.01, ***p < 0.001 as determined by Student's t test.
(E) Bar graph of the top 15 GO terms obtained from alternative splicing analysis from IFE keratinocytes.
(F) Bar graph of the top 15 GO terms obtained from alternative splicing analysis from ORS keratinocytes.
(G) Venn diagram of alternative splicing targets from P2 IFE and ORS cells.
(H) Bar graph of the significantly enriched GO terms obtained from overlapping genes displaying alternatively spliced exons or junction sites from IFE and ORS keratinocytes.
(I) Pie chart of the ratio of alternatively used exons and junction sites of genes within the GO term “cell cycle.”