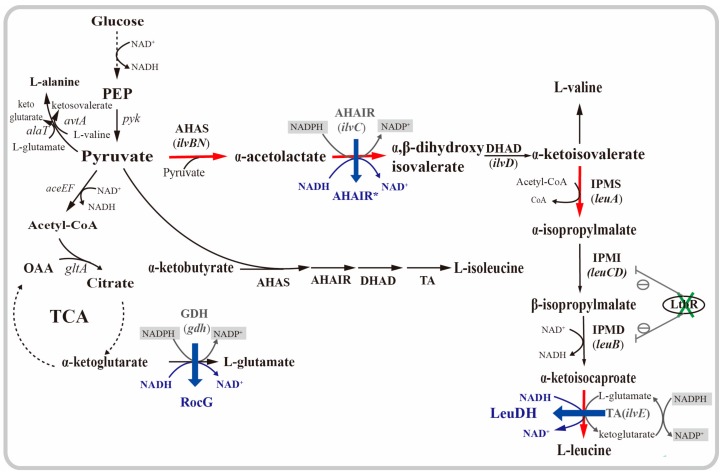
Figure 1.
The l-leucine biosynthesis pathway of Corynebacterium glutamicum and the metabolic engineering steps performed in this study. Enzymes and corresponding genes are shown. An encircled minus indicates repression of gene expression (solid lines). Deletion of the gene encoding LtbR is indicated by green “×”. Red thick arrows indicate increased metabolic fluxes. Introduction of enzymatic reactions are highlighted in blue. NADPH and NADP+ are highlighted in grey shaded boxes. Abbreviations: AHAS—acetotydroxyacid synthetase; AHAIR—acetohydroxyacid isomeroreductase; DHAD—dihydroxyacid dehydratase; TA—branched-chain amino acid transaminase; IPMS—α-isopropylmalate synthase; IPMI—α-isopropylmalate isomerase; IPMD—β-isopropylmalate dehydrogenase; LeuDH—leucine dehydrogenase; PEP—phosphoenolpyruvate; OAA—oxaloacetic acid; TCA—tricarboxylic acid cycle; GDH and RocG—glutamate dehydrogenase.