Abstract
Background
The high mortality of patients with non-small cell lung cancer (NSCLC) emphasizes the necessity of identifying a robust and reliable prognostic signature for NSCLC patients. This study aimed to identify and validate a prognostic signature for the prediction of both disease-free survival (DFS) and overall survival (OS) of NSCLC patients by integrating multiple datasets.
Methods
We firstly downloaded three independent datasets under the accessing number of GSE31210, GSE37745 and GSE50081, and then performed an univariate regression analysis to identify the candidate prognostic genes from each dataset, and identified the gene signature by overlapping the candidates. Then, we built a prognostic model to predict DFS and OS using a risk score method. Kaplan–Meier curve with log-rank test was used to determine the prognostic significance. Univariate and multivariate Cox proportional hazard regression models were implemented to evaluate the influences of various variables on DFS and OS. The robustness of the prognostic gene signature was evaluated by re-sampling tests based on the combined GEO dataset (GSE31210, GSE37745 and GSE50081). Furthermore, a The Cancer Genome Atlas (TCGA)-NSCLC cohort was utilized to validate the prediction power of the gene signature. Finally, the correlation of the risk score of the gene signature and the Gene set variation analysis (GSVA) score of cancer hallmark gene sets was investigated.
Results
We identified and validated a six-gene prognostic signature in this study. This prognostic signature stratified NSCLC patients into the low-risk and high-risk groups. Multivariate regression and stratification analyses demonstrated that the six-gene signature was an independent predictive factor for both DFS and OS when adjusting for other clinical factors. Re-sampling analysis implicated that this six-gene signature for predicting prognosis of NSCLC patients is robust. Moreover, the risk score of the gene signature is correlated with the GSVA score of 7 cancer hallmark gene sets.
Conclusion
This study provided a robust and reliable gene signature that had significant implications in the prediction of both DFS and OS of NSCLC patients, and may provide more effective treatment strategies and personalized therapies.
Electronic supplementary material
The online version of this article (10.1186/s12967-019-1899-y) contains supplementary material, which is available to authorized users.
Keywords: Non-small cell lung cancer, Disease-free survival, Overall survival, Risk score, Prognostic signature
Background
Lung cancer is the leading cause of cancer death worldwide, and non-small cell lung cancer (NSCLC) composes the majority (approximately 85%) of all lung cancers [1, 2]. Despite advances in treatment strategies, the high mortality rate for lung cancer patients has not considerably declined, due to the late diagnosis of the disease [3]. The major clinical determinants of NSCLC prognosis include tumor extension, performance status and histological type [4, 5]. However, various disease outcomes have been identified in patients with similar clinical and pathological features, suggesting that the current clinical prognostic factors used may be insufficient to consistently predict individual clinical outcomes [6]. This emphasizes the necessity of identifying robust and reliable prognostic markers with higher sensitivity and accuracy in NSCLC.
Transcriptome profiling has widely been used to characterize prognostic signatures in patients with lung cancer, and has generated a number of candidate biomarkers with potential clinical values [7–9]. However, the suggested signatures lack consistency among studies and provide limited prognostic information, partially due to the limited sample size and technical factors. Moreover, NSCLC is a highly heterogeneous disease, thus it is critical to identify a reliable signature that can define patients who are at a high-risk to develop disease recurrence. To this end, integrating the results from multiple studies holds promise for more robust prognostic signatures. In addition, most investigations used overall survival (OS) rather than tumor recurrence as an end point [9–11]. Disease-free survival (DFS) is defined as the interval from surgery to the first diagnosis of any type of relapse or death, and is used as a possible alternative for OS.
Therefore, we attempted to identify and validate a robust and reliable prognostic signature for DFS and OS prediction by integrating multiple datasets of NSCLC patients. In the present study, we revealed a six-gene signature with a reliable prognostic value in NSCLC, which might complement conventional clinical prognostic factors, and further provide more effective therapeutic interventions and personalized therapies for NSCLC patients.
Methods
Patient data
Gene expression data and corresponding clinical information data of NSCLC patients were obtained from the publicly available database Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). Three independent datasets were collected in this study, under the accessing number of GSE31210 [12, 13], GSE37745 [14] and GSE50081 [15]. These gene expression data were generated using the same chip platform Affymetrix HG-U133 Plus 2.0 platform. GSE31210 consisted of a total of 226 lung adenocarcinoma cases. GSE37745 included 196 NSCLC cases, including 106 adenocarcinoma, 24 large cell carcinoma, and 66 squamouscarcinoma. GSE50081 included 181 NSCLC cases, including 127 adenocarcinoma, 7 large cell carcinoma, 43 squamous carcinoma, and 4 adenosquamous carcinoma. A total of 603 cases were enrolled for OS analysis, including 226 patients from GSE31210, 196 patients from GSE37745, and 181 patients from GSE50081. For DFS analysis, a total of 499 patients were finally included, including 226 patients from GSE31210, 96 patients from GSE37745, and 177 patients from GSE50081. The details of the patients’ clinical information in each dataset are described in Tables 1 and 2. All microarray data were normalized using robust multi-array average (RMA) and microarray Suite 5 (MAS5) methods, and log2-scale transformed in this study.
Table 1.
Univariate and multivariate Cox regression analysis of the gene signature and disease-free survival of NSCLC patients
| Variables | Patients (N) |
Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| GSE31210 | ||||||
| Age | ≤ 65/> 65 | 176/50 | 1.89 (1.12–3.18) | 1.70E−02 | 2.36 (1.37–4.04) | 1.83E−03 |
| Gender | Female/male | 121/105 | 1.27 (0.78–2.07) | 3.38E−01 | ||
| ALK fusion | +/− | 215/11 | 0.74 (0.18–3.02) | 6.73E−01 | ||
| EGFR mutation | +/− | 99/127 | 0.60 (0.37–0.98) | 4.29E−02 | 1.98 (0.88–4.47) | 1.00E−01 |
| KRAS mutation | +/− | 206/20 | 0.98 (0.42–2.26) | 9.56E−01 | ||
| Triple negative | No/yes | 158/68 | 1.87 (1.14–3.08) | 1.39E−02 | 2.91 (1.30–6.54) | 9.71E−03 |
| Myc copy | Low/high | 207/17 | 1.04 (0.42–2.59) | 9.33E−01 | ||
| Stage | I/II | 168/58 | 3.16 (1.92–5.21) | 6.09E−06 | 2.77 (1.63–4.71) | 1.62E−04 |
| Smoking | No/yes | 115/111 | 1.33 (0.82–2.18) | 2.52E−01 | ||
| Risk score | Low/high | 123/103 | 3.26 (1.92–5.53) | 1.24E−05 | 3.10 (1.77–5.41) | 7.17E−05 |
| GSE37745 | ||||||
| Age | ≤ 65/> 65 | 45/51 | 0.89 (0.50–1.55) | 6.70E−01 | ||
| Gender | Female/male | 46/50 | 1.01 (0.58–1.78) | 9.60E−01 | ||
| Stage | I/II | 63/18 | 1.13 (0.54–2.38) | 7.45E−01 | ||
| Stage | I/III | 63/14 | 1.78 (0.81–3.91) | 1.48E−01 | ||
| Stage | I/IV | 63/1 | 5.71 (0.75–43.63) | 9.32E−02 | ||
| Histological type | Adeno/large | 53/13 | 1.05 (0.46–2.42) | 9.08E−01 | ||
| Histological type | Adeno/squamous | 53/30 | 1.12 (0.60–2.10) | 7.17E−01 | ||
| WHO performance status | 0/1 | 54/32 | 2.21 (1.21–4.03) | 1.02E−02 | 2.43 (1.32–4.47) | 4.31E−03 |
| WHO performance status | 0/2 | 54/6 | 1.04 (0.25–4.42) | 9.52E−01 | 1.12 (0.26–4.75) | 8.77E−01 |
| WHO performance status | 0/3 | 54/4 | 1.66 (0.50–5.53) | 4.08E−01 | 1.62 (0.49–5.38) | 4.34E−01 |
| Risk score | Low/high | 46/50 | 2.31 (1.27–4.21) | 6.08E−03 | 2.49 (1.36–4.55) | 3.10E−03 |
| GSE50081 | ||||||
| Age | ≤ 65/> 65 | 59/118 | 1.24 (0.69–2.24) | 4.77E−01 | ||
| Gender | Female/male | 81/96 | 1.68 (0.95–2.99) | 7.59E−02 | ||
| Stage | I/II | 124/53 | 1.87 (1.06–3.28) | 3.03E−02 | 1.71 (0.97–3.01) | 6.30E−02 |
| Histological type | Adeno/large | 124/7 | 1.04 (0.25–4.31) | 9.59E−01 | ||
| Histological type | Adeno/squamous | 124/42 | 0.73 (0.10–5.36) | 7.61E−01 | ||
| Histological type | Adeno/other | 124/4 | 0.74 (0.38–1.45) | 3.79E−01 | ||
| Smoking | No/yes | 24/133 | 0.70 (0.35–1.40) | 3.14E−01 | ||
| Risk score | Low/high | 83/94 | 2.42 (1.33–4.43) | 4.02E−03 | 2.30 (1.26–4.22) | 6.97E−03 |
| All patients | ||||||
| Age | ≤ 65/> 65 | 280/219 | 1.36 (1.00–1.84) | 5.04E−02 | ||
| Gender | Female/male | 248/251 | 1.31 (0.96–1.78) | 8.51E−02 | ||
| Stage | I/II | 355/129 | 2.05 (1.47–2.85) | 2.02E−05 | 1.72 (1.23–2.40) | 1.50E−03 |
| Stage | I/III | 355/14 | 3.45 (1.68–7.11) | 7.74E−04 | 2.62 (1.27–5.43) | 9.41E−03 |
| Stage | I/IV | 355/1 | 22.26 (3.01–164.44) | 2.36E−03 | 14.79 (2.00–109.7) | 8.40E−03 |
| Histological type | Adeno/large | 403/20 | 1.48 (0.75–2.93) | 2.61E−01 | ||
| Histological type | Adeno/squamous | 403/72 | 0.76 (0.11–5.44) | 7.85E−01 | ||
| Histological type | Adeno/other | 403/4 | 1.17 (0.77–1.78) | 4.52E−01 | ||
| Risk score | Low/high | 252/247 | 2.7 (1.94–3.76) | 4.16E−09 | 2.39 (1.70–3.35) | 4.47E−07 |
HR: hazard ratio; CI: confidence interval; Adeno: adenocarcinoma; Large: large cell carcinoma; Squamous: squamous cell carcinoma
Table 2.
Univariate and multivariate Cox regression analysis of the gene signature and overall survival of NSCLC patients
| Variables | Patients (N) |
Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| GSE31210 | ||||||
| Age | ≤ 65/> 65 | 176/50 | 2.58 (1.31–5.08) | 5.99E−03 | 3.51 (1.72–7.15) | 5.44E−04 |
| Gender | Female/male | 121/105 | 1.52 (0.78–2.96) | 2.19E−01 | ||
| ALK fusion | +/− | 215/11 | 1.49 (0.36–6.24) | 5.82E−01 | ||
| EGFR mutation | +/− | 99/127 | 0.47 (0.24–0.93) | 2.96E−02 | 1.87 (0.65–5.37) | 2.47E−01 |
| KRAS mutation | +/− | 206/20 | 0.87 (0.27–2.85) | 8.17E−01 | ||
| Triple negative | No/yes | 158/68 | 2.19 (1.12–4.26) | 2.12E−02 | 2.95 (1.06–8.19) | 3.76E−02 |
| Myc copy | Low/high | 207/17 | 0.7 (0.17–2.9) | 6.18E−01 | ||
| Stage | I/II | 168/58 | 4.23 (2.17–8.24) | 2.17E−05 | 3.45 (1.72–6.93) | 5.06E−04 |
| Smoking | No/yes | 115/111 | 1.64 (0.84–3.2) | 1.50E−01 | ||
| Risk score | Low/high | 129/97 | 6.14 (2.68–14.07) | 1.84E−05 | 5.47 (2.3–12.99) | 1.21E−04 |
| GSE37745 | ||||||
| Age | ≤ 65/> 65 | 102/94 | 1.35 (0.98–1.88) | 6.88E−02 | ||
| Gender | Female/male | 89/107 | 1.1 (0.79–1.52) | 5.85E−01 | ||
| Stage | I/II | 130/35 | 1.22 (0.79–1.87) | 3.66E−01 | 1.14 (0.73–1.77) | 5.68E−01 |
| Stage | I/III | 130/27 | 1.86 (1.19–2.93) | 6.88E−03 | 1.63 (1.02–2.63) | 4.27E−02 |
| Stage | I/IV | 130/4 | 1.31 (0.42–4.15) | 6.43E−01 | 1.97 (0.61–6.35) | 2.57E−01 |
| Histological type | Adeno/large | 106/24 | 0.89 (0.52–1.53) | 6.75E−01 | ||
| Histological type | Adeno/squamous | 106/66 | 1.26 (0.88–1.79) | 2.05E−01 | ||
| WHO performance status | 0/1 | 105/75 | 1.91 (1.35–2.7) | 2.38E−04 | 1.96 (1.39–2.79) | 1.53E−04 |
| WHO performance status | 0/2 | 105/12 | 1.95 (1.01–3.79) | 4.83E−02 | 1.81 (0.92–3.57) | 8.66E−02 |
| WHO performance status | 0/3 | 105/4 | 1.13 (0.36–3.61) | 8.32E−01 | 1.09 (0.34–3.54) | 8.80E−01 |
| Risk score | Low/high | 97/99 | 1.56 (1.12–2.16) | 8.31E−03 | 1.44 (1.02–2.04) | 3.80E−02 |
| GSE50081 | ||||||
| Age | ≤ 65/> 65 | 59/122 | 1.56 (0.93–2.61) | 9.04E−02 | ||
| Gender | Female/male | 83/98 | 1.93 (1.19–3.14) | 7.80E−03 | 2 (1.23–3.26) | 5.47E−03 |
| Stage | I/II | 127/54 | 1.69 (1.05–2.72) | 3.10E−02 | 1.63 (1.01–2.63) | 4.71E−02 |
| Histological type | Adeno/large | 127/7 | 1.71 (0.62–4.74) | 3.01E−01 | ||
| Histological type | Adeno/squamous | 127/43 | 1.84 (0.57–5.92) | 3.03E−01 | ||
| Histological type | Adeno/other | 127/4 | 0.8 (0.46–1.39) | 4.35E−01 | ||
| Smoking | No/yes | 24/136 | 1.39 (0.66–2.92) | 3.89E−01 | ||
| Risk score | Low/high | 79/102 | 2.21 (1.34–3.64) | 1.81E−03 | 2.09 (1.27–3.45) | 3.91E−03 |
| All patients | ||||||
| Age | ≤ 65/> 65 | 337/266 | 1.92 (1.5–2.46) | 2.48E−07 | 1.83 (1.42–2.35) | 2.94E−06 |
| Gender | Female/male | 293/310 | 1.45 (1.13–1.86) | 3.79E−03 | 1.27 (0.98–1.64) | 7.14E−02 |
| Stage | I/II | 425/147 | 1.67 (1.26–2.21) | 3.43E−04 | 1.5 (1.13–2) | 5.31E−03 |
| Stage | I/III | 425/27 | 3.6 (2.34–5.56) | 7.02E−09 | 2.79 (1.79–4.36) | 6.58E−06 |
| Stage | I/IV | 425/4 | 2.53 (0.81–7.93) | 1.12E−01 | 3.23 (1.02–10.23) | 4.58E−02 |
| Histological type | Adeno/large | 459/31 | 1.76 (1.1–2.81) | 1.83E−02 | 1.51 (0.94–2.43) | 9.16E−02 |
| Histological type | Adeno/squamous | 459/109 | 2.29 (0.73–7.18) | 1.56E−01 | 1.92 (0.6–6.1) | 2.70E−01 |
| Histological type | Adeno/other | 459/4 | 1.85 (1.39–2.45) | 2.09E−05 | 1.27 (0.94–1.72) | 1.18E−01 |
| Risk score | Low/high | 305/298 | 2.11 (1.63–2.72) | 1.03E−08 | 1.65 (1.26–2.18) | 3.32E−04 |
HR: hazard ratio; CI: confidence interval; Adeno: adenocarcinoma; Large: large cell carcinoma; Squamous: squamous cell carcinoma
The genomic data and clinical information of NSCLC patients in The Cancer Genome Atlas (TCGA) were obtained from the University of California Santa Cruz Xenabrowser (UCSC Xena, http://xena.ucsc.edu/) [16]. This cohort has 761 NSCLC patients with the corresponding gene expression data (read count) and clinical information (including survival data).
Prognostic gene signature screening
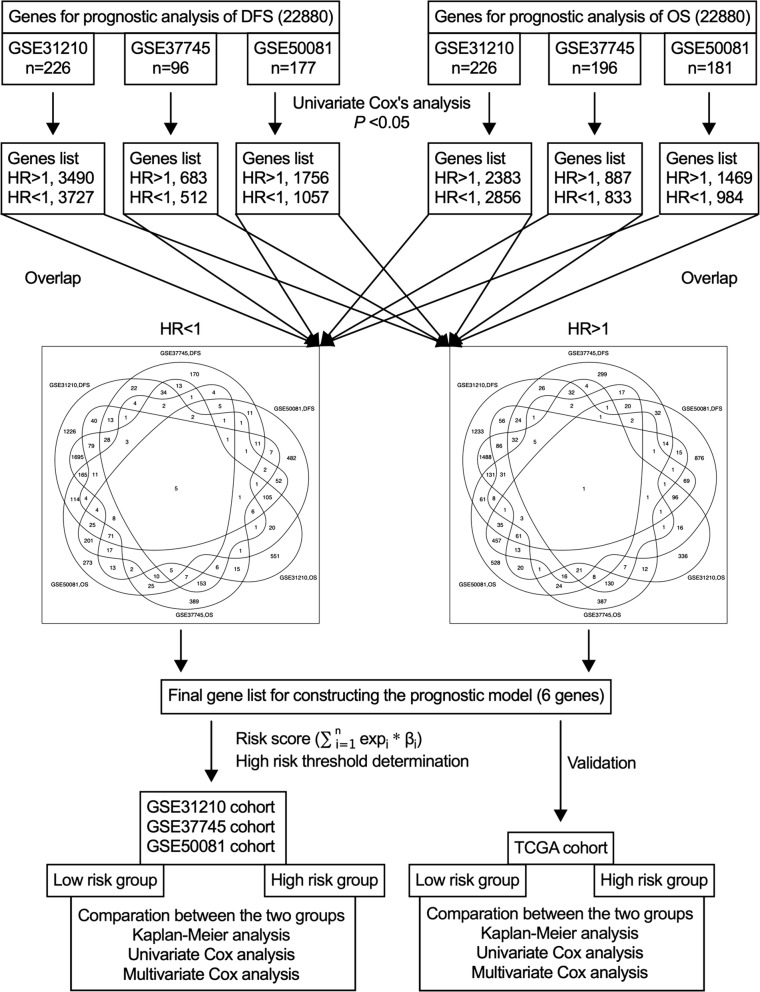
In this study, we firstly screened candidate prognostic genes from each cohort, and selected the common ones for constructing the prognostic gene signature. Then, the prognostic value of the signature was validated using each cohort. The flow diagram of this study is illustrated in Fig. 1. An univariate Cox proportional hazard regression model was implemented to determine the association of gene expression with DFS and OS in each cohort. Genes under a cutoff value of P < 0.05 were defined as candidate genes related to OS and DFS, and the common genes among three datasets were selected to construct the prognostic signature. Hazard ratio (HR) from the univariate Cox regression analysis was used to determine the protective (HR < 1) and risky genes (HR > 1).
Fig. 1.
Flow diagram of this study
Then, a risk score was established for each patient by calculating the expression values of the selected genes weighted by regression coefficients in the univariate Cox regression analysis. The formula used was as follows:
Where n is the number of selected genes, expi is the expression level of gene i, and βi represents the regression coefficient of gene i. Subsequently, the risk score was dichotomized at the median value, and patient whose risk score was greater than the median value was divided into a high-risk group, otherwise into alow-risk group.
Evaluation of the robustness of prognostic gene signature by re-sampling tests
The data from three GEO datasets (GSE31210 [12, 13], GSE37745 [14] and GSE50081 [15]) were combined. Then a subset that contained 70% samples of the combined GEO dataset (re-sampling) was randomly selected and used to determine the prediction power of the gene signature for DFS and OS of the NSCLC patients. This re-sampling test was repeated 100 times.
Association of prognostic gene signature and cancer hallmarks
A total of 50 hallmark gene sets which are currently recognized were downloaded from the molecular signature database (MSigDB, http://software.broadinstitute.org/gsea/msigdb). Next, gene set variation analysis (GSVA) package and its ssGSEA method (http://www.bioconductor.org) were implemented for these 50 hallmark gene sets to further obtain the GSVA scores of each gene set for each sample in the combined GEO datasets [17]. The GSVA score devotes the degree of absolute enrichment of a gene set in each sample. After that, Pearson’s correlation analysis was performed to investigate whether the GSVA score of the members of the given gene set was correlated with the risk score. The correlation coefficients (R), confidence interval (CI) and P values were calculated.
Statistical analysis
The DFS and OS were calculated using Kaplan–Meier curves, and the statistical difference was determined by log-rank test. Influences of various variables on DFS and OS were evaluated by univariate and multivariate Cox proportional hazard regression models. HR and the 95% CI were generated using Cox proportional hazards models. The receiver operating characteristic (ROC) curve analysis was carried out to compare the predictive accuracy of the gene signature. A P value < 0.05 was set as the statistically as the significant difference.
Results
Identification of a six-gene prognostic signature
We firstly identified survival-related genes using univariate Cox regression analysis in each dataset. Under the cut-off threshold of Cox P < 0.05, 7217 genes in GSE31210, 1195 genes in GSE37745 and 2813 genes in GSE50081 were identified as candidate predictive genes that presented close association with DFS. Similarly, 2539 genes in GSE31210, 1720 genes in GSE37745 and 2453 genes in GSE50081 were identified to be involved in OS. By overlapping these candidate genes among three datasets, a set of 6 common genes was screened finally, including one risky gene (HR > 1) and 5 protective genes (HR < 1). The general information of these 6 genes is displayed in Table 3.
Table 3.
Overall information of the 6 genes for constructing the prognostic signature
| Gene stable ID | Gene name | Gene type | Chromosome | Gene start (bp) | Gene end (bp) |
|---|---|---|---|---|---|
| ENSG00000152527 | PLEKHH2 | Protein coding | 2 | 43,637,273 | 43,767,987 |
| ENSG00000136003 | ISCU | Protein coding | 12 | 108,562,582 | 108,569,384 |
| ENSG00000079101 | CLUL1 | Protein coding | 18 | 596,988 | 650,334 |
| ENSG00000101938 | CHRDL1 | Protein coding | X | 110,673,856 | 110,795,819 |
| ENSG00000124374 | PAIP2B | Protein coding | 2 | 71,182,739 | 71,227,083 |
| ENSG00000163814 | CDCP1 | Protein coding | 3 | 45,082,278 | 45,146,422 |
The six-gene signature predicts survival of NSCLC patients
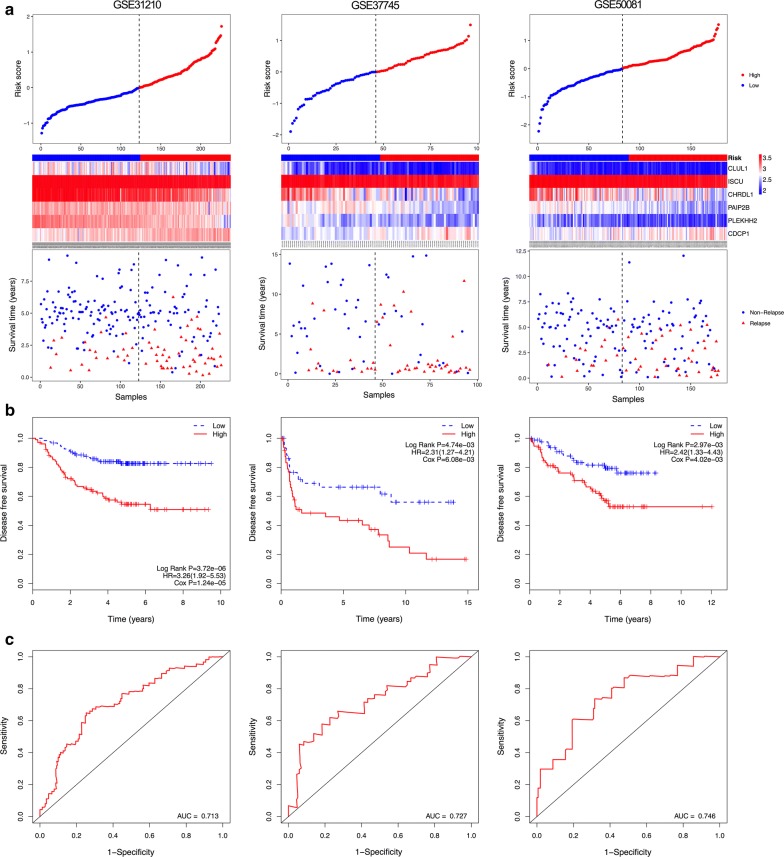
According to the gene expression and regression coefficients of the 6 genes, a prognostic model was developed to predict prognosis using a risk score method. In the prognostic model, each patient was endowed a risk score. Using the median risk score value as the cut-off point, patients in each dataset were classified into low-risk and high-risk groups. The DFS prediction power of the six-gene signature for patients in each dataset is displayed in Fig. 2. The distribution of gene risk scores, gene expression levels, and patients’ relapse status in each dataset are shown in Fig. 2a.
Fig. 2.
Correlation between the six-gene signature and the disease-free survival (DFS) of patients in three datasets. a The distribution of risk scores, gene expression levels and patient relapse status. b Kaplan–Meier curves of DFS of the low- and high-risk groups. c ROC curve for the 5-year survival prediction by the six-gene signature. The black dotted line in a represents the median risk score cut-off dividing patients into low- and high-risk groups
Kaplan–Meier curves showed that patients in the high-risk groups presented significantly shorter DFS than those in the low-risk groups (GSE31210: HR = 3.26, 95% CI 1.92–5.53, P < 0.05; GSE37745: HR = 2.31, 95% CI 1.27–4.21, P < 0.05; GSE50081: HR = 2.42, 95% CI 1.33–4.43, P < 0.05) (Fig. 2b). Furthermore, a time-dependent ROC curve was performed to evaluate the sensitivity and specificity of the six-gene signature for DFS prediction. Notably, the six-gene signature achieved AUC values of 0.713 in GSE31210, 0.727 in GSE37745 and 0.746 in GSE50081 (Fig. 2c), suggesting a substantially effective performance for DFS prediction.
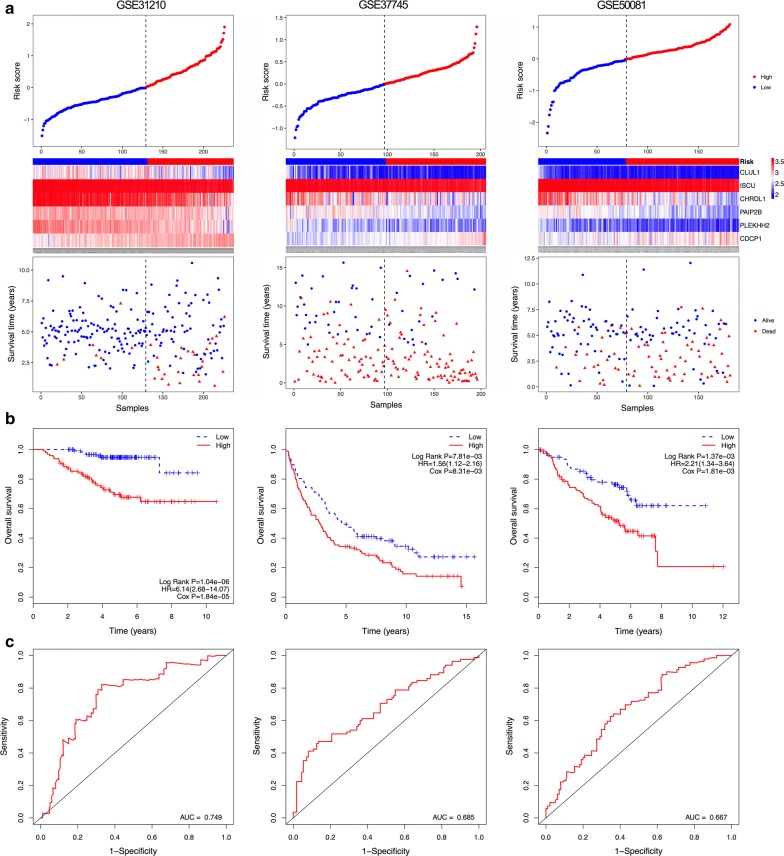
The OS prediction value of the six-gene signature for patients in each dataset is shown in Fig. 3. Figure 3a illustrates the distribution of gene risk scores, gene expression levels and patients’ survival status in each dataset. Consistent with our previous finding, patients in the high-risk groups had significantly shorter OS when compared with those in the low-risk groups (GSE31210: HR = 6.14, 95% CI 2.68–14.07, P < 0.05; GSE37745: HR = 1.56, 95% CI 1.12–2.16, P < 0.05; GSE50081: HR = 2.21, 95% CI 1.34–3.64, P < 0.05) (Fig. 3b). Patients with high risk scores tended to have poorer clinical outcomes compared with those with low risk scores. In addition, the time-dependent ROC curve was implemented to measure the sensitivity and specificity of the six-gene signature for OS prediction in each dataset. Markedly, the signature achieved AUC values of 0.749, 0.685 and 0.667 in GSE31210, GSE37745 and GSE50081, respectively (Fig. 3c), implying a high OS prediction performance.
Fig. 3.
Correlation between the six-gene signature and overall survival (OS) of patients in three datasets. a The distribution of risk scores, gene expression levels and patient survival status. b Kaplan–Meier curves of OS of low- and high-risk groups. c ROC curve for the 5-year survival prediction by the six-gene signature. The black dotted line in a represents the median risk score cut-off dividing patients into low- and high-risk groups
The six-gene prognostic signature is robust
Previous study has demonstrated that tumor heterogeneity limits the generation of robust prognostic biomarker [18]. Thus, we conducted re-sampling tests for validation of the robustness of the prognostic gene signature. As shown in Additional file 1: Table S1, we found that in all the random 100 model validations of prediction power of the gene signature for OS by re-sampling, the P values were less than 0.0001 in each univariate Cox and Kaplan–Meier analysis. Notably, the six-gene signature achieved the AUC values of more than 0.650 for 1, 2, 3, 4, and 5-year OS in the combined GEO datasets, demonstrating a high OS prediction performance. Similarly, among all the random 100 model validations of prediction power of the gene signature for DFS, the P values were less than 0.0001 in each univariate Cox and Kaplan–Meier analysis (Additional file 2: Table S2). Moreover, the six-gene signature obtained the AUC values of more than 0.610 for 1, 2, 3, 4, and 5-year DFS in the combined GEO datasets (Additional file 2: Table S2), which implicates that this signature has an effective performance for DFS prediction. Overall, these results suggest this six-gene signature for predicting prognosis of NSCLC patients is robust.
The six-gene signature is an independent prognostic factor
Here, we performed univariate and multivariate Cox regression models in these three datasets. The six-gene risk score and other clinicopathological factors, including age, gender, stage, histological type, gene mutation, smoking and performance status were used as covariates. The association between these factors and DFS is shown in Table 1. Univariate regression analysis indicated that age, EGFR mutation, triple negative status, disease stage and risk score were significantly associated with the DFS of NSCLC patients in GSE31210; WHO performance status and risk score were significantly associated with the DFS of patients in GSE37745; and stage and risk score were related to the DFS of patients in GSE50081. In the entire cohort, stage and risk score were identified to have significant correlation with the DFS of NSCLC patients. Moreover, in order to determine whether the six-gene signature was independent of other clinical factors, we performed a multivariate regression analysis, and found a significant correlation of the six-gene signature with DFS in three datasets (GSE31210: HR = 3.10, 95% CI 1.77–5.41, P = 7.17E−05; GSE37745: HR = 2.49, 95% CI 1.36–4.55, P = 3.10E−03; GSE50081: HR = 2.30, 95% CI 1.26–4.22, P = 6.97E−03) and the entire cohort (HR = 2.39, 95% CI = 1.70−3.35, P = 4.47E−07),after adjusting for other clinical factors. The result indicated that the six-gene risk score was an independent adverse DFS factor for NSCLC patients.
The correlation of risk score and other clinicopathological factors with the OS of NSCLC patients is shown in Table 2. We performed an univariate regression analysis to determine the correlation between these factors and OS. Our results indicated that age, EGFR mutation, triple negative status, stage and risk score were OS prognostic factors for NSCLC patients in GSE31210; stage I/III, WHO performance status and risk score were significantly related to OS of patients in GSE37745; and age, stage and risk score were OS prognostic factors for patients in GSE50081. In the entire cohort, age, gender, stage and risk score were correlated with OS of NSCLC patients. Subsequent multivariate regression analysis indicated that the six-gene signature was an independent OS prognostic factor in three datasets (GSE31210: HR = 5.47, 95% CI 2.30–12.99, P = 1.21E−04; GSE37745: HR = 1.44, 95% CI 1.02–2.04, P = 3.80E−02; GSE50081: HR = 2.09, 95% CI 1.27–3.45, P = 3.91E−03) and entire cohort (HR = 1.65, 95% CI 1.26–2.18, P = 3.32E−04), after adjusting for other clinical factors. Taken together, our data show that the six-gene risk score was an independent adverse prognostic factor for both DFS and OS of NSCLC patients.
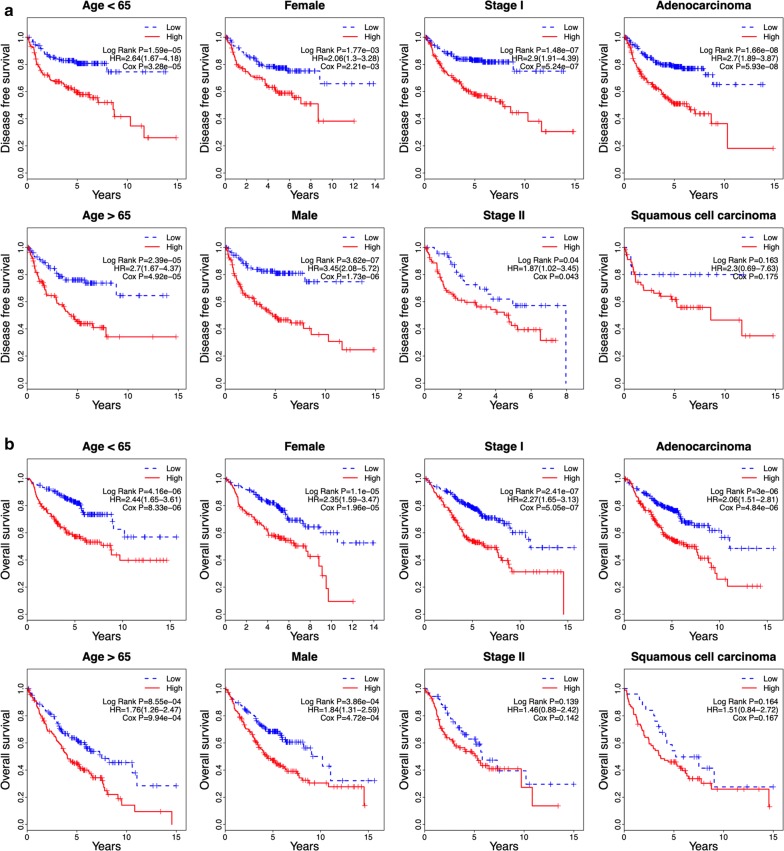
Furthermore, we performed a data stratification analysis on the entire cohort. These patients (499 patients for DFS and 603 for OS) were factitiously stratified based on their clinical parameters, such as age (≤ 65/> 65), gender (female/male), stage (I/II) and histological type (adenocarcinoma/squamous carcinoma). Because of the small sample size, patients in stage III and IV, and patients with large cell cancer were removed from the stratification analysis. The results showed that the six-gene risk score remained the ability of predicting the DFS and OS within each stratum. In Fig. 4a, the results of stratification analysis showed that high-risk patients in each stratum of age, gender and early stage had significantly shorter DFS than low-risk patients (P < 0.05). For patients with adenocarcinoma, high-risk patients showed significantly shorter DFS than low-risk patients (P < 0.05), while there was no significant difference between high-risk and low-risk patients for patients with squamous carcinoma, might due to the small sample size of patients with squamous carcinoma. In Figure 4b, the results of stratification analysis indicated that high-risk patients in each stratum presented significantly poorer OS than low-risk patients (P < 0.05), except for patients in stage II and patients with squamous carcinoma. Taken together, our findings suggested that the six-gene signature was independent of other clinical features for DFS and OS prediction in NSCLC patients.
Fig. 4.
Kaplan–Meier analysis of DFS and OS for NSCLC patients stratified by age, gender, stage and histological type
Further validation of the six-gene signature using another independent dataset
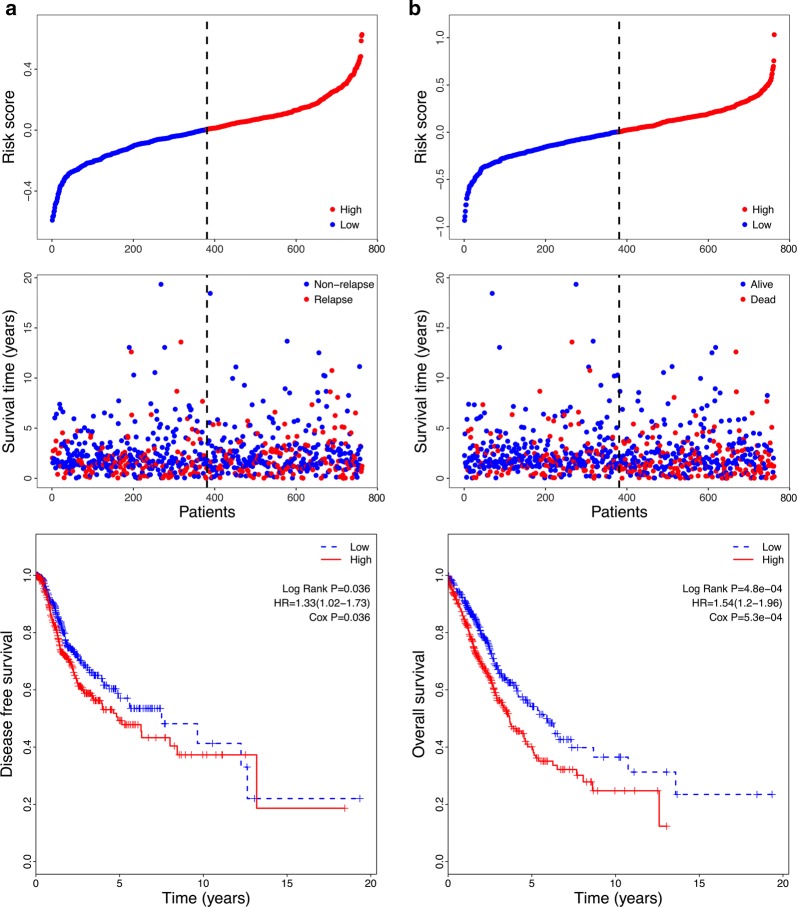
To investigate the reliability of the six-gene signature, another independent dataset from the TCGA was used for further validation. The risk score of each sample in this dataset was calculated, and the samples were then classified into low- and high-risk groups using the median risk score value as the cut-off point (Fig. 5a, b). Kaplan–Meier and univariate Cox regression analysis exhibited that the patients in the high-risk group had obviously shorter DFS than those in the low-risk groups (HR = 1.33, 95% CI 1.02–1.73, P < 0.05) (Fig. 5a and Table 4). Similarly, patients in the high-risk group presented remarkably shorter OS compared to those in the low-risk groups (HR = 1.54, 95% CI 1.20–1.96, P < 0.05) (Fig. 5b and Table 4). Univariate Cox regression analysis also indicated that stage II, stage III and squamous histologic type were poor prognostic factors for prediction of DFS, and stage II, stage III and stage IV were poor prognostic factors for prediction of OS (Table 4). However, subsequent multivariate regression analysis indicated that only stage II, stage III and risk score were independent prognostic factors for prediction of both DFS and OS (P < 0.05). These results suggest that the six-gene signature is valid and reliable across datasets and platforms.
Fig. 5.
Correlation between the six-gene signature and DFS/OS of patients in the TCGA dataset. a The distribution of risk scores, patient relapse status, and Kaplan–Meier curves of DFS of low- and high-risk groups. b The distribution of risk scores, patient survival condition, and Kaplan-Meier curves of OS of low- and high-risk groups
Table 4.
Univariate and multivariate Cox regression analysis of the gene signature and survival of NSCLC patients in TCGA cohort
| Variables | Group | Patients (N) |
Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| DFS | ||||||
| Age | ≤ 67/> 67 | 383/378 | 0.97 (0.75–1.26) | 8.30E−01 | 1.11 (0.85–1.45) | 4.30E−01 |
| Gender | Female/male | 326/435 | 0.96 (0.74–1.25) | 7.60E−01 | 1.08 (0.81–1.43) | 6.00E−01 |
| Stage | I/II | 410/220 | 1.64 (1.21–2.22) | 1.30E−03 | 1.74 (1.28–2.37) | 3.60E−04 |
| Stage | I/III | 410/109 | 2.16 (1.51–3.08) | 2.20E−05 | 2.25 (1.57–3.22) | 9.30E−06 |
| Stage | I/IV | 410/22 | 1.76 (0.82–3.80) | 1.50E−01 | 1.56 (0.71–3.40) | 2.70E−01 |
| Histological type | Adeno/squamous | 395/366 | 0.65 (0.50–0.85) | 1.60E−03 | 0.58 (0.43–0.77) | 1.60E−04 |
| Risk score | Low/high | 380/381 | 1.33 (1.02–1.73) | 3.70E−02 | 1.40 (1.08–1.83) | 1.30E−02 |
| OS | ||||||
| Age | ≤ 67/> 67 | 383/378 | 1.25 (0.98–1.58) | 7.10E−02 | 1.34 (1.05–1.70) | 1.90E−02 |
| Gender | Female/male | 326/435 | 1.22 (0.96–1.57) | 1.10E−01 | 1.19 (0.92–1.53) | 2.00E−01 |
| Stage | I/II | 410/220 | 1.51 (1.14–2.00) | 4.30E−03 | 1.45 (1.09–1.93) | 1.10E−02 |
| Stage | I/III | 410/109 | 2.07 (1.50–2.85) | 9.60E−06 | 2.04 (1.47–2.83) | 1.80E−05 |
| Stage | I/IV | 410/22 | 2.77 (1.59–4.83) | 3.20E−04 | 2.83 (1.60–5.00) | 3.40E−04 |
| Histological type | Adeno/squamous | 395/366 | 1.13 (0.89–1.44) | 3.20E−01 | 0.93 (0.71–1.22) | 5.90E−01 |
| Risk score | Low/high | 380/381 | 1.54 (1.20–1.96) | 5.50E−04 | 1.48 (1.14–1.91) | 3.30E−03 |
HR: hazard ratio; CI: confidence interval; Adeno: adenocarcinoma; Squamous: squamous cell carcinoma
The six-gene signature is association with several hallmarks
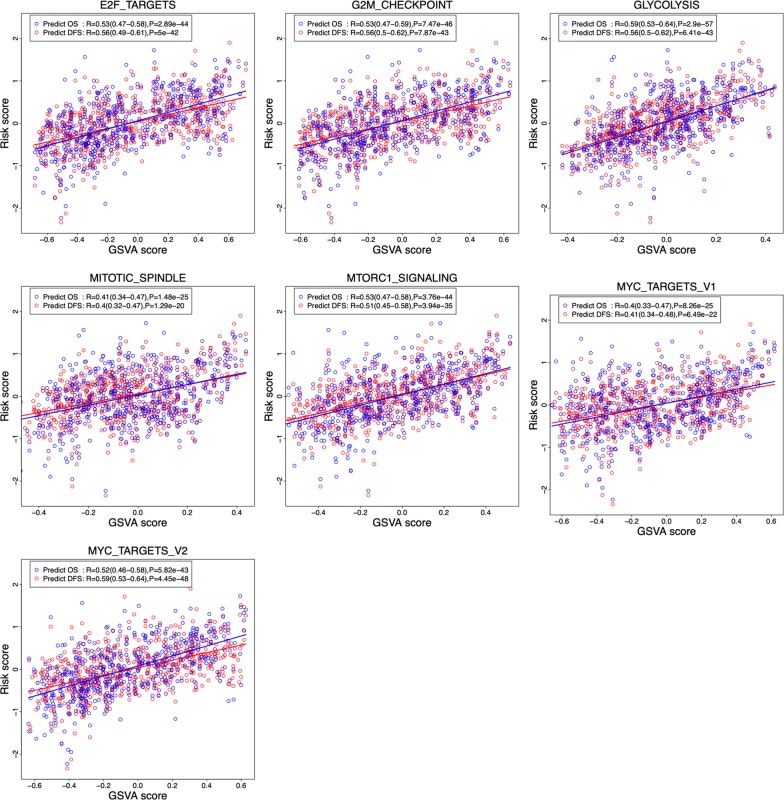
To identify the six-gene signature associated biological processes, the correlation of the risk score of the gene signature for predicting DFS/OS and the GSVA score of cancer hallmark gene sets was investigated. As shown in Fig. 6, a total of 7 hallmark gene sets (E2F_TARGETS, G2M_CHECKPOINT, GLYCOLYSIS, MITOTIC_SPINDLE, MTORC1-SIGNALING, MYC-TARGETS-V1, MYC-TARGETS-V2) were identified to be correlated with risk score [correlation coefficients (R) is higher than or equal to 0.4; P < 0.0001]. Interestingly, these biological processes and the risk score displayed the same trend, suggesting that activation of theses hallmarks might participate in the process of tumor progression and affect the survival of the patients with NSCLC.
Fig. 6.
Association between the risk score of the six-gene prognosis signature and the GSVA score of 7 cancer hallmark gene sets
Discussion
Numerous reports have indicated that disturbed gene expression may be implicated in various aspects of tumor, including tumorigenesis, progression and prognosis [19–21]. Some genes have been considered as prospective biomarkers to predict prognosis in NSCLC patients [14, 22, 23]. However, several concerns limit their prognostic and predicative power, such as inadequate samples, lack of DFS prediction, and lack of effective validation. In this study, we developed and validated a novel prognostic six-gene signature that was found to be significantly associated with both the DFS and OS of NSCLC patients. Our results revealed that this classifier could successfully identify high-risk and low-risk NSCLC patients with significant differences in both DFS and OS. In addition, the prognostic value of the six-gene signature was verified in three GEO datasets and an independent TCGA dataset, suggesting the reproducibility and reliability of the six-gene signature for both DFS and OS prediction in NSCLC.
The clinical prognostic factors in NSCLC include stage, age, gender and performance status [6, 24]. Our study showed that stage and age were significantly correlated with both the DFS and OS of patients in GSE31210; performance status was significantly associated with DFS, and stage and performance status were related to OS of patients in GSE37745; stage and gender were significantly involved in OS of patients in GSE50081. In the entire patient cohort, stage was identified as an independent prognostic factor for DFS, and age and stage were associated with OS of NSCLC patients. Furthermore, we performed a stratification analysis on the entire cohort, and found that the prognostic power of the six-gene signature was independent of age, gender and stage. Interestingly, Birim et al. [24] indicated that non-squamous cell histology was a risk factor for postoperative outcome in NSCLC. Our study showed that histological type had no significant association with either DFS or OS in NSCLC. While stratification analysis indicated that high-risk group had significantly shorter DFS and OS than low-risk group for patient with adenocarcinoma but not squamous carcinoma.
Currently, tumor stage has been broadly utilized as a strong indicator of survival in NSCLC [25]. However, the current staging system is far from accurate in the aspect of survival prediction at the individual level [26]. As expected in our study, univariate and multivariate analysis showed that stage II, stage III, and stage IV were significantly associated with OS and DFS in the entire GEO cohort. However, stage II, and stage III were found to be the independent prognostic factors for prediction of both DFS and OS in the TCGA database. As documented, age is a main indicator of patient survival, and younger patients are tended to survive longer than the older ones [27–29]. Nevertheless, age alone is not a survival indicator for cancer patients because older patients are less likely to receive adjuvant therapy [30]. Multivariate analysis showed that age was significantly associated with OS in the entire GEO cohort and the TCGA cohort, but there was no correlation between DFS and age in these two cohorts. Thus, compared to age and stage, the risk score was a more reliable prognostic factor for NSCLC patients.
Several gene mutations have been revealed to be associated with the pathogenesis of NSCLC, such as EGFR and KRAS mutations [31]. Notably, EGFR or KRAS mutated lung cancer accounts for a significant subgroup of NSCLC, especially in adenocarcinoma [32, 33]. Markedly, targeting EGFR mutations has changed the therapeutic paradigm in NSCLC patients harboring EGFR mutations [34]. Over the last decade, multiple EGFR tyrosine kinase inhibitors (TKIs) have been developed to target mutated EGFR, and have achieved a better survival in patients with EGFR mutations than in those with the wild type [35]. Whereas, KRAS mutations predict a worse prognosis among NSCLC patients treated by chemotherapy and EGFR-TKIs [36, 37]. In the present study, univariate analysis indicated that EGFR mutations, but not KRAS mutations, were correlated with both DFS and OS, whereas multivariate analysis indicated that EGFR mutation status did not act as an independent prognostic factor.
In this study, we developed a prognostic six-gene signature for both DFS and OS prediction in NSCLC. Most of these genes have not been well characterized in tumor biology, except for CDCP1. CDCP1, also known as CUB domain-containing protein 1, is a transmembrane glycoprotein, whose phosphorylation is linked with the progression and metastasis of several cancers [38, 39]. In addition, blocking of CDCP1 has been shown to be a potential mode for therapeutic intervention against metastatic disease [38, 40]. Chiu et al. [41] showed that the ADAM9 metalloprotease enhanced CDCP1 expression via activating EGFR signaling pathways in advanced lung cancer disease. Ikeda et al. [42] revealed that CDCP1 expression was an independent prognostic factor for both OS and DFS, and could be used as an useful marker for survival prediction of patients with lung adenocarcinoma. Our study combined these 6 genes into a single panel, and established its prognostic value in both DFS and OS in NSCLC.
As reported, the complexity of cancer can be decreased and presented by a few cancer hallmarks that enable cancer cell proliferation and metastasis. These hallmarks can offer a framework to understand the cancer diversity. Hence, we focused on detecting the association of prognostic gene signature and cancer hallmarks [43]. Studies have revealed that DNA replication, cell cycle, DNA damage repair, apoptosis, chromosome and gene instability, energy supply play important roles in cancer progression [44–47]. Coincidentally, the GSVA results demonstrated that the six-gene signature was remarkably connected with these biological processes. Specifically, E2F targets have been demonstrated to participate in DNA replication, cell cycle, DNA damage repair, apoptosis [48]. G2M checkpoint, mitotic spindle, and MYC targets have been reported to contribute to the instability of chromosome and gene [49–51]. Utilization of glycolysis-related metabolic pathway has been implicated to provide ATP as a main source of energy supply for cancers [52]. Moreover, mTORC1 signaling has been suggested to be activated in human fibrolamellar liver carcinoma [53]. Demonstrated here, the results of present study demonstrated that the six-gene signature correlated with several cancer-progression associated biological processes which supported the DFS/OS predictive ability of the signature. Significantly, the correlation analysis in our study showed that patients having these activated biological processes tended to have adverse outcomes. Thus, this further confirmed that our six-gene signature used for predicting prognosis was reasonable and reliable.
In this work, some limitations need to be acknowledged. First, a few clinical characters presented an unbalanced distribution, such as an overwhelming majority of patients in stage I/II and presenting a histological adenocarcinoma type. Thus, the robustness of the six-gene signature requires further validation in large-scale prospective investments. Second, most of the genes identified here are rarely reported in the academic literature, and there are no experimental data regarding the identified signature, thus more evidence is needed to elucidate the inherent correlation between the six-gene signature and the prognosis of NSCLC patients. Despite these drawbacks, our results demonstrate valuable information on the importance and significance of the six-gene signature in both DFS and OS prediction in NSCLC.
Conclusions
In this study, we developed an innovative six-gene prognostic signature for both DFS and OS prediction in NSCLC patients. The six-gene signature was an independent prognostic factor, and might complement clinicopathological factors and facilitate the personalized treatment of NSCLC patients. Large-scale prospective investments should be applied for further assessment of the robustness of this signature in future studies.
Additional files
Additional file 1: Table S1. Validating the prediction power of the gene signature for OS in the combined GEO dataset by re-sampling analysis.
Additional file 2: Table S2. Validating the prediction power of the gene signature for DSF in the combined GEO dataset by re-sampling analysis.
Acknowledgements
Not applicable.
Authors’ contributions
Conceived and designed the study: SZ. Analyzed the data: SZ, MW, HZ, JD, and AC. Contributed to reviewing/revising: JW, JD, and JW. Wrote the paper: SZ. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81472820, 81773255 and 81700037), Natural Science Foundation of Jiangsu Province of China (BK20171098), Fundamental Research Funds for the Central Universities (14380336/1-2), and Six talent peaks project in Jiangsu Province.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuguang Zuo, Min Wei and Hailin Zhang contributed equally to this work
Contributor Information
Shuguang Zuo, Email: zuosg@icloud.com.
Min Wei, Email: MG1835023@smail.nju.edu.cn.
Hailin Zhang, Email: DG1635008@smail.nju.edu.cn.
Anxian Chen, Email: chenanxian2017@smail.nju.edu.cn.
Junhua Wu, Email: wujunhua@nju.edu.cn.
Jiwu Wei, Email: wjw@nju.edu.cn.
Jie Dong, Email: dongjie0805@163.com.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Marijon H, Bouyon A, Vignot S, Besse B. Prognostic and predictive factors in lung cancer. Bull Cancer. 2009;96(4):391–404. doi: 10.1684/bdc.2009.0779. [DOI] [PubMed] [Google Scholar]
- 5.Cuyun Carter G, Barrett AM, Kaye JA, Liepa AM, Winfree KB, John WJ. A comprehensive review of nongenetic prognostic and predictive factors influencing the heterogeneity of outcomes in advanced non-small-cell lung cancer. Cancer Manag Res. 2014;6:437–449. doi: 10.2147/CMAR.S63603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brundage MD, Davies D, Mackillop WJ. Prognostic factors in non-small cell lung cancer: a decade of progress. Chest. 2002;122(3):1037–1057. doi: 10.1378/chest.122.3.1037. [DOI] [PubMed] [Google Scholar]
- 7.Sanfiorenzo C, Ilie MI, Belaid A, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS ONE. 2013;8(1):e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Y, Wang L, Liu P, Yang P, You M. Gene-expression signature predicts postoperative recurrence in stage I non-small cell lung cancer patients. PLoS ONE. 2012;7(1):e30880. doi: 10.1371/journal.pone.0030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 10.Lin T, Fu Y, Zhang X, et al. A seven-long noncoding RNA signature predicts overall survival for patients with early stage non-small cell lung cancer. Aging (Albany NY) 2018;10(9):2356–2366. doi: 10.18632/aging.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Tang H, Sun Z, et al. Network-based approach identified cell cycle genes as predictor of overall survival in lung adenocarcinoma patients. Lung Cancer. 2013;80(1):91–98. doi: 10.1016/j.lungcan.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi M, Yamaguchi R, Nakata A, et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS ONE. 2012;7(9):e43923. doi: 10.1371/journal.pone.0043923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botling J, Edlund K, Lohr M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19(1):194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 15.Der SD, Sykes J, Pintilie M, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9(1):59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 16.Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci Rep. 2013;3(10):2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Lenferink AE, Deng Y, et al. Corrigendum: identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 2010;1(4):34. doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthaios D, Hountis P, Karakitsos P, Bouros D, Kakolyris S. H2AX a promising biomarker for lung cancer: a review. Cancer Invest. 2013;31(9):582–599. doi: 10.3109/07357907.2013.849721. [DOI] [PubMed] [Google Scholar]
- 20.Borczuk AC, Toonkel RL, Powell CA. Genomics of lung cancer. Proc Am Thorac Soc. 2009;6(2):152–158. doi: 10.1513/pats.200807-076LC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu CQ, Tsao MS. Prognostic markers in lung cancer: is it ready for prime time? Transl Lung Cancer Res. 2014;3(3):149–158. doi: 10.3978/j.issn.2218-6751.2014.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang S, Wang Z. EGFR mutations as a prognostic and predictive marker in non-small-cell lung cancer. Drug Des Devel Ther. 2014;8:1595–1611. doi: 10.2147/DDDT.S69690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farhat FS, Tfayli A, Fakhruddin N, et al. Expression, prognostic and predictive impact of VEGF and bFGF in non-small cell lung cancer. Crit Rev Oncol Hematol. 2012;84(2):149–160. doi: 10.1016/j.critrevonc.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Birim O, Kappetein AP, van Klaveren RJ, Bogers AJ. Prognostic factors in non-small cell lung cancer surgery. Eur J Surg Oncol. 2006;32(1):12–23. doi: 10.1016/j.ejso.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Woodard GA, Jones KD, Jablons DM. Lung cancer staging and prognosis. Cancer Treat Res. 2016;170:47–75. doi: 10.1007/978-3-319-40389-2_3. [DOI] [PubMed] [Google Scholar]
- 26.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2(8):706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y, Scheck AC, Cloughesy TF, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan ER, Norman A, Laing K, Seal MD. Treatment and outcomes for glioblastoma in elderly compared with non-elderly patients: a population-based study. Curr Oncol. 2017;24(2):e92–e98. doi: 10.3747/co.24.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris G, Jayamanne D, Wheeler H, et al. Survival outcomes of elderly patients with glioblastoma multiforme in their 75th year or older treated with adjuvant therapy. Int J Radiat Oncol Biol Phys. 2017;98(4):802–810. doi: 10.1016/j.ijrobp.2017.02.028. [DOI] [PubMed] [Google Scholar]
- 30.Gately L, Collins A, Murphy M, Dowling A. Age alone is not a predictor for survival in glioblastoma. J Neurooncol. 2016;129(3):479–485. doi: 10.1007/s11060-016-2194-x. [DOI] [PubMed] [Google Scholar]
- 31.Guan JL, Zhong WZ, An SJ, et al. KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol. 2013;20(4):1381–1388. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 32.Vincenten JP, Smit EF, Grunberg K, et al. Is the current diagnostic algorithm reliable for selecting cases for EGFR- and KRAS-mutation analysis in lung cancer? Lung Cancer. 2015;89(1):19–26. doi: 10.1016/j.lungcan.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Fan G, Zhang K, Ding J, Li J. Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(20):33922–33932. doi: 10.18632/oncotarget.15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loong HH, Kwan SS, Mok TS, Lau YM. Therapeutic strategies in EGFR mutant non-small cell lung cancer. Curr Treat Options Oncol. 2018;19(11):58. doi: 10.1007/s11864-018-0570-9. [DOI] [PubMed] [Google Scholar]
- 35.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 36.Nygaard AD, Garm Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79(3):312–317. doi: 10.1016/j.lungcan.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Martin P, Leighl NB, Tsao MS, Shepherd FA. KRAS mutations as prognostic and predictive markers in non-small cell lung cancer. J Thorac Oncol. 2013;8(5):530–542. doi: 10.1097/JTO.0b013e318283d958. [DOI] [PubMed] [Google Scholar]
- 38.Kollmorgen G, Niederfellner G, Lifke A, et al. Antibody mediated CDCP1 degradation as mode of action for cancer targeted therapy. Mol Oncol. 2013;7(6):1142–1151. doi: 10.1016/j.molonc.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Ong SE, Badu-Nkansah K, Schindler J, White FM, Hynes RO. CUB-domain-containing protein 1 (CDCP1) activates Src to promote melanoma metastasis. Proc Natl Acad Sci U S A. 2011;108(4):1379–1384. doi: 10.1073/pnas.1017228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casar B, He Y, Iconomou M, Hooper JD, Quigley JP, Deryugina EI. Blocking of CDCP1 cleavage in vivo prevents Akt-dependent survival and inhibits metastatic colonization through PARP1-mediated apoptosis of cancer cells. Oncogene. 2012;31(35):3924–3938. doi: 10.1038/onc.2011.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu KL, Lin YS, Kuo TT, et al. ADAM9 enhances CDCP1 by inhibiting miR-1 through EGFR signaling activation in lung cancer metastasis. Oncotarget. 2017;8(29):47365–47378. doi: 10.18632/oncotarget.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda J, Oda T, Inoue M, et al. Expression of CUB domain containing protein (CDCP1) is correlated with prognosis and survival of patients with adenocarcinoma of lung. Cancer Sci. 2009;100(3):429–433. doi: 10.1111/j.1349-7006.2008.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang E, Zaman N, McGee S, Milanese JS, Masoudi-Nejad A, O’Connor-McCourt M. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Cárcer GD, Venkateswaran SV, Salgueiro L, et al. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat Commun. 2018;9(1):3012. doi: 10.1038/s41467-018-05429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2015;16(1):35. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 46.Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med. 2016;6(3):a026104. doi: 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaube B, Bhat MK. AMPK, a key regulator of metabolic|[sol]|energy homeostasis and mitochondrial biogenesis in cancer cells. Cell Death Dis. 2016;7(1):e2044. doi: 10.1038/cddis.2015.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Krempler A, Deckbar D, Jeggo PA, Löbrich M. An imperfect G2M checkpoint contributes to chromosome instability following irradiation of S and G2 phase cells. Cell Cycle. 2007;6(14):1682–1686. doi: 10.4161/cc.6.14.4480. [DOI] [PubMed] [Google Scholar]
- 50.Gulluni F, Martini M, Santis MCD, et al. Mitotic spindle assembly and genomic stability in breast cancer require PI3K-C2α scaffolding function. Cancer Cell. 2017;32(4):444. doi: 10.1016/j.ccell.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Kumari A, Folk WP, Sakamuro D. The dual roles of MYC in genomic instability and cancer chemoresistance. Genes. 2017;8(6):158. doi: 10.3390/genes8060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelicano H, Martin DS, Xu R-H, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2015;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 53.Riehle KJ, Yeh MM, Yu JJ, et al. mTORC1 and FGFR1 signaling in fibrolamellar hepatocellular carcinoma. Mod Pathol. 2015;28(1):103–110. doi: 10.1038/modpathol.2014.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Validating the prediction power of the gene signature for OS in the combined GEO dataset by re-sampling analysis.
Additional file 2: Table S2. Validating the prediction power of the gene signature for DSF in the combined GEO dataset by re-sampling analysis.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.