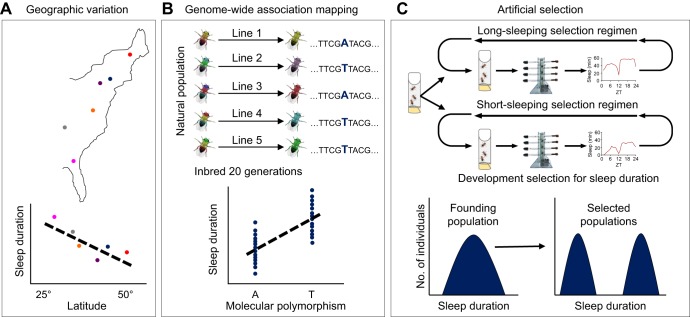
Fig. 2.
Approaches for studying the contributions of natural variation on sleep regulation in fruit flies. (A) Drosophila melanogaster from different geographical regions have been tested for sleep. Increased sleep duration is associated with proximity to the equator (Svetec et al., 2015). (B) Genome-wide association mapping studies have been performed using the Drosophila Genetics Research Panel. Individual inbred fly lines originating from wild-caught populations are tested for sleep and phenotypes are associated with genetic variation (Harbison et al., 2013). A and T represent single nucleotide genomic differences between individual lines. (C) Artificial selection of outbred lines for long- or short-sleep phenotypes, resulting in dramatic shifts in sleep duration. Flies were grown in vials and then selected as long or short sleepers using Drosophila Activity Monitors (DAMs) (Seugnet et al., 2009 and Masek et al., 2014).