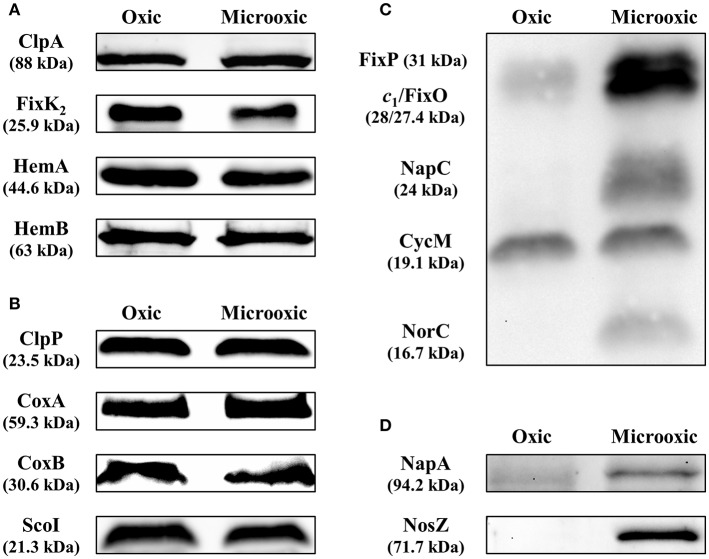
Figure 5.
Validation of protein abundance for genes subject to post-transcriptional regulation determined by heme-staining and western blot analyses. Steady-state levels of ClpA, FixK2, HemA, and HemB (A), targets subject to post-transcriptional control, were analyzed by western blot. ClpP, CoxA, CoxB, and ScoI, proteins with constant accumulated levels, were included as controls (B). Up-regulation of the membrane-bound FixO and FixP proteins and the periplasmic, soluble NapA and NosZ proteins were monitored by heme-staining (C) or western blot (D). The membrane-bound cytochrome CycM was used as reference in the heme-staining experiments in (C) as the protein levels remained constant in both oxic and microoxic conditions. Crude extracts, soluble and membrane fractions were isolated from B. diazoefficiens cells cultivated oxically and microoxically. 10–40 μg of cytosolic (ClpA, ClpP, HemA, HemB, NapA, NosZ), membrane (CoxB, ScoI), or crude extract (CoxA, FixK2) fractions were loaded in the gel in the western blot experiments (identical amount of protein extracted from cells grown oxically and microoxically for each validated target). Twenty-five microgram membranes were loaded in the gel for the heme-staining analyses. Apparent molecular masses of the proteins are shown on the left. Note that as described previously (Loferer et al., 1993), the CoxA protein migrated at ≈45 kDa instead of the corresponding predicted mass of 59.3 kDa. Shown are representative results of different experiments carried out with at least three independent biological replicates. A detailed description of the methodology is described in Supplementary Table S5.