Figure 1.
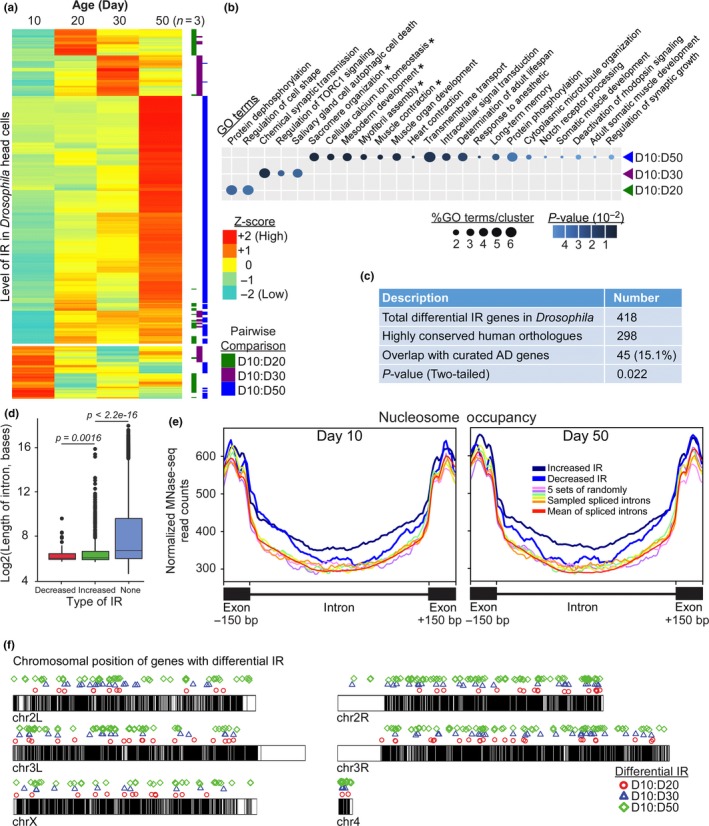
Differential intron retentions (IR) mark different stages of aging in Drosophila. (a) Expression heatmap of the differentially retained introns across various ages as represented by Z‐score. Pairwise comparison of the two age‐groups is highlighted by the color bar on the right. (b) Gene ontology analysis of the genes with differential IR in aging fly heads (p‐value <0.05, Fisher's exact test; *p‐value <0.05, Fisher's exact test with Benjamini–Hochberg correction). (c) The overlap between highly conserved human orthologues of fly differential IR genes and curated AD genes from DisGeNET. The p‐value was determined by two‐tailed chi‐square test with Yates’ continuity correction. (d) Boxplot for the length distribution of the various types of introns where D10 flies were used as the reference to determine the differential decreased or increased IR during aging. “None” refers to non‐retained, spliced introns (p < 0.05, Welch's t test). (e) Normalized MNase‐seq read counts across different types of introns in Day 10 and 50 fly heads showed that nucleosome occupancy over differentially retained introns was significantly different from the spliced introns (p < 0.05, Wilcoxon rank sum test). The increased or decreased IR referred to the differentially retained introns at Day 50 with respect to Day 10. Five sets of spliced introns with similar expression level were randomly picked as control. (f) Ideogram displaying the genome‐wide distribution of the differential IR genes in fly
