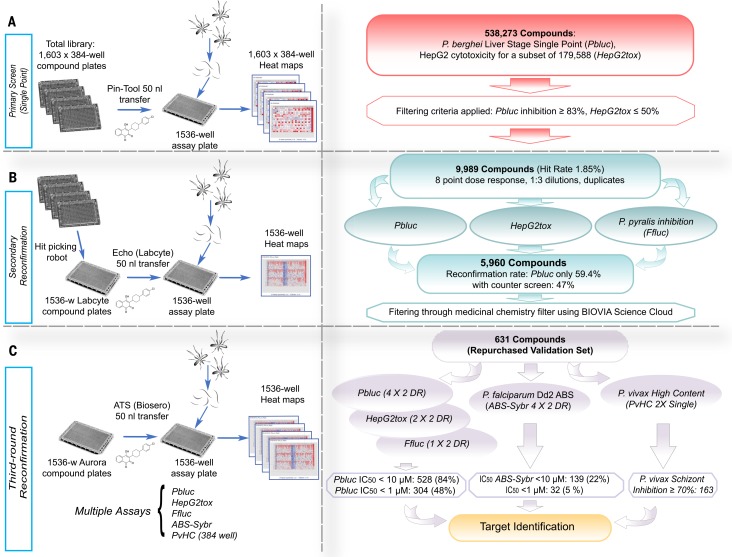
Fig. 1.
Screen workflow. (A) The Charles River library (538,273 compounds) was plated into 1603 384-well plates. For the primary Pbluc single-point screen, compounds from four of the 384-well plates were transferred with a pin-tool instrument (50 nl per well) into 1536-well assay plates containing seeded HepG2 cells (3 × 103 cells per well).The next day, P. berghei luciferase–expressing sporozoites were freshly prepared from infected Anopheles stephensi mosquitoes, and ~1000 sporozoites in a 5 ml volume were added to each well. After 48 hours, P. berghei–Luc growth within hepatocytes was measured with bioluminescence. (B) To prepare source plates for the first round of reconfirmation (second round of screening), the 9989 hit compounds were transferred from the original 384-well library with an automated hit-picking system and serially diluted into eight points (1:3 dilutions) for dose-response screening in duplicate. The hit compounds (50 nl per well) in serial dilutions were acoustically transferred into assay wells containing HepG2 cells for Pbluc and HepG2tox assays. In addition, biochemical recombinant luciferase inhibition assay (Ffluc) were also performed. (C) For final reconfirmation (third round), 631 compounds prepared from re-sourced powders and were serially diluted (10 points, 1:3 dilution) and plated into Aurora 1536- well compound plates. Compounds (50 nl per well) were acoustically transferred into 1536-well assay plates. Multiple dose-respose assays such as Pbluc, HepG2tox, Ffluc, and ABS-Sybr were performed to determine IC50 in the third round of screening. A P. vivax liver schizont formation high-content assay in single-point (2X) was also performed.