Abstract
Membrane mimetics are essential to study the structure, dynamics and function of membrane-associated proteins by biophysical and biochemical approaches. Among various membrane mimetics that have been developed and demonstrated for studies on membrane proteins, lipid nanodiscs are the latest developments in the field and are increasingly used for various applications. While lipid-nanodiscs can be formed using an amphipathic membrane scaffold protein (MSP), peptide, or synthetic polymer, the synthetic polymer based nanodiscs exhibit unique advantages because of the ability to functionalize them for various applications. In addition to the use of synthetic polymers to extract membrane proteins directly from the cell membranes, recent advances in the development of polymers used for nanodiscs formation are attracting new attention to the field of nanodiscs technology. Here we review the developments of novel polymer modifications that overcome the current limitations and enhance the applications of polymer based nanodiscs to a wider variety of biophysical techniques used to study membrane proteins. A summary of the functionalization of poly(Styrene-co-Maleic Acid), SMA, polymers developed by our research and their advantages are also covered in this review article.
1. Introduction
1.1. Membrane mimetics are essential to functionally reconstitute membrane proteins
Biological lipid membranes are a vital component of any cell. Lipid membranes provide functional barriers between subcellular compartments and between the cell and its environment. Membrane proteins are essential parts of the cellular membrane that facilitates many of the crucial cellar functions required for life.[1–5] Approximately 30% of the human genome is dedicated to encoding membrane proteins [6] and they represent ~60% of all drug targets [7]. However, only ~3% of all current protein structures reported in the Protein Data Bank (PDB) [8] are membrane proteins. The lack of membrane protein structural information limits our current understanding of cellular membrane biology [9] and significantly hinders the process of modern drug design [10]. Numerous challenges have plagued the study of functional membrane protein structures. A crucial challenge in the structural biology investigation of membrane proteins is their functional stability. Most membrane-associated proteins are insoluble in aqueous solvents. One common method for membrane protein solubilization is the use of detergent micelles [11, 12]. Detergents, small amphiphilic molecules that consist of long hydrophobic chains and a polar head group, have been the most commonly used method for extracting, isolating, and purifying membrane proteins from their native cellular environment [13, 14]. Membrane protein solubilization by detergents consists of an undesirable perturbation of the native lipid bilayer and an association of the detergent hydrophobic tails with the hydrophobic transmembrane region of the membrane protein. This extraction and micelle formation commonly interferes with membrane protein folding, causing a loss in membrane protein native functionality [15–19]. This major drawback greatly hinders the study of membrane proteins prompting the need for better membrane mimetics.
To circumvent this problem a common membrane mimetic used in membrane protein research is the liposome [9, 20]. Liposomes are vesicles that consist of lipids that spontaneously assembled into a bilayer. These are advantageous over detergents as the liposomal bilayer closely resembles the cellular membrane environment as compared to a detergent micelle [20]. The major drawback of liposomes as a membrane mimetic is their relative instability, their limited use in various biophysical characterization techniques, and the need for detergent inclusion in the extraction process of membrane proteins [21]. Another common membrane mimetic used to study membrane proteins are bicelles [22–25], which are disc-shaped phospholipid bilayers surrounded by a rim containing short chained detergent molecules. The advantages of bicelles over liposomes are that bicelles have no membrane curvature, higher stability, and size tunability. Particularly, the size tunability is achieved by changing the ratio of lipid to detergent (q-ratio) [26]. By controlling the q-ratio, bicelles have been shown to align in the presence of a magnetic field at larger sizes (q>2.5). These large bicelles are anisotropic as they do not tumble fast enough in the NMR time scale. [27] Bicelles have also been used for solution NMR studies when formed at lower sizes (q < 0.5; also known as isotropic bicelles) [28, 29]. While advantageous over liposomes, bicelles still have the crucial problem of including detergents during the reconstitution of membrane proteins. The detergent molecules present in bicelles undergo diffusion from the rim to the planar lipid bilayer and also present in the form of toroidal pores within the planar lipid bilayer of the bicelles [30]. The ability of the detergent molecule to diffuse into the lipid bilayer can denature an embedded protein [9]. To overcome these challenges researchers have developed nanodiscs to better simulate a native-like membrane environment for membrane protein research [31].
1.2. Curvature-free lipid nanodiscs enabled structural studies of membrane proteins
Introduced by Sligar and co-workers in 2002, nanodiscs are non-covalent nanoparticles that consist of a disc shaped lipid bilayer stabilized by an amphiphilic membrane scaffold protein (MSP) [32] that was inspired from high density lipid particles [33]. MSP nanodiscs can be made by mixing MSPs with detergent-solubilized protein and lipids [31]. Scaffold protein nanodiscs have been very successful in the study of several membrane proteins [33–39] and amyloid proteins [40, 41]. The curvature-free nanodiscs have been demonstrated to be highly valuable to understand amyloid aggregation mechanism and to trap amyloid oligomers for high-resolution NMR based structural studies [40]. However, the process of incorporating proteins into MSP nanodiscs still requires the use of detergents during the reconstitution process [31]. MSP based nanodiscs also introduce interfering spectral properties during the study of reconstituted membrane proteins. Even though very recent studies reported the possibility of increasing the size of MSP-based nanodiscs [37], which are otherwise small (typically <15 nm diameter), there are difficulties in reconstituting large-size membrane proteins or protein-protein complexes. Some of these difficulties have been overcome using short amphipathic peptides engineered from the MSP protein [42, 43]. These peptides have been shown to self-assemble with lipids to form nanodiscs and enable the reconstitution of protein-protein complexes. These nanodiscs undergo collision and exchange lipid contents as demonstrated by a recent study using high-speed AFM and 31P NMR experiments [44–46]. The peptide-based nanodiscs are useful for structural studies of membrane proteins using solution NMR [42], SAXS [47], solid-state NMR [48] and also used for potential cancer immunotherapy [49, 50]. While peptide-based nanodiscs are increasingly used, their interference with biophysical studies of the embedded protein of interest and other potential undesired effects for in-vivo applications are inherent limitations for further biological and biomedical applications. Therefore, to overcome these limitations, there is significant interest in the development of different types of amphipathic molecules that can form nanodiscs.
2. Polymer based lipid-nanodiscs
Poly(Styrene-co-Maleic Acid), SMA, is an amphiphilic polymer that is obtained by hydrolyzation of the poly(Styrene-co-Maleic Anhydride), SMAnh [51–53]. SMA based nanodiscs are a promising technology that can be used for isolation, purification, structural and functional characterization of membrane proteins [52, 54]. In 2009 SMA-lipid particles (SMALPs) were first reported to have the ability to form nanodiscs [55]. SMA also allows the direct extraction of membrane proteins without removing them from their native lipid membrane environment and, most importantly, this process is accomplished without the use of destabilizing detergents [54, 56, 57]. Most of the current SMA research has involved investigating several different molecular weights and styrene to maleic anhydride ratios, leading to a plethora of different polymers available commercially [58, 59]. The advantages over micelles, liposomes, bicelles, and protein based nanodiscs allow polymer nanodiscs to be applied to a wider variety of membrane protein studies.
Polymer nanodiscs in the form of SMALPs have been shown to have significant downsides to their use however. The original SMA polymers (~9.5 kDa) used in SMALPs did not allow for any significant size control over the nanodiscs [53], also all SMA based polymer nanodiscs are unstable to conditions that require low pH or the presence of divalent metal ions [51, 54]. These limitations have restricted the applications of SMA based nanodiscs and do not allow for the active study of membrane proteins whose function require a low pH or require the presence of divalent metal ions such as Ca2+ or Mg2+ [60–62]. The instability of SMA is due to the presence of carboxylic acids as the hydrophilic portion of the polymer. SMA is only soluble in water at a high pH that allows for the formation of carboxylates, however carboxylates also strongly chelate with divalent metal ions, leading to SMAs instability [51, 54]. The anhydride unit of SMAnh copolymers can be functionalized by amines via a nucleophilic ring opening reaction[63]. SMAnh can be customized in several different ways and our research has focused on the hydrophilic functionalization of SMA, while also imparting size control by employing a lower molecular weight SMA as our starting material.
SMA polymers have been very successful in the reconstitution of membrane proteins into nanodiscs [52, 54, 64–71]. SMA polymers have the unique advantage over other membrane mimetics in that SMA has been shown to directly extract membrane proteins from the cellular membrane [52, 54, 56]. SMA polymer nanodiscs have been shown to form in a variety of lipids with a size range of 10–15 nm diameter [53, 72]. A variety of biophysical studies reported the characterization of SMA based polymer nanodiscs [73–75]. NMR experiments have been used to show the formation of nanodiscs and to probe lipid dynamics [76, 77]. Due to the lack of relative size control the current NMR applications of polymer nanodiscs are limited to solution NMR. In order to expand the NMR applications of polymer nanodiscs to both solution and solid-state NMR, more control over nanodiscs size is needed [22, 39, 78]. Our hypothesis is that a lower molecular weight polymer would allow for size control of nanodiscs is based on the knowledge gained from the molecular weight (MW) difference between high MW MSPs, which do not allow for size control, and low MW peptide based nanodiscs which have been demonstrated to enable size control [35, 42]. Our lab has been focused on using low molecular weight polymers to achieve the necessary nanodiscs size control for various biophysical and structural biology studies (Figure 1).
Figure 1. SMA derivatives that form lipid nanodiscs.
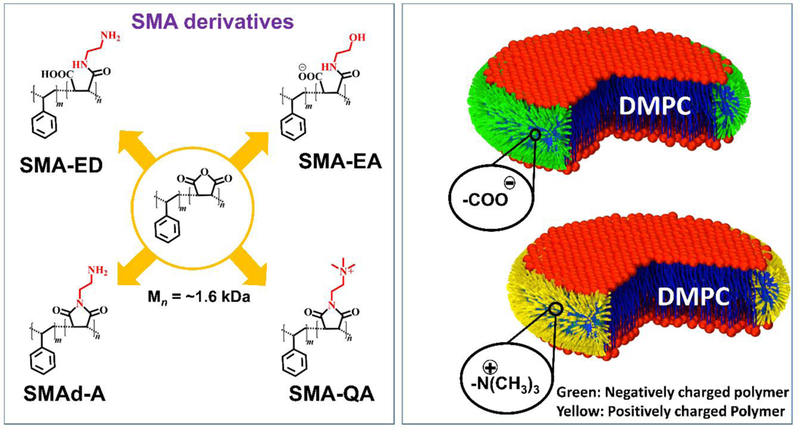
Hydrophilic functionalization to enhance polymer nanodisc features and applications. Molecular structures of SMA based polymer derivatives used to form lipid-nanodiscs (left panel) and illustrations of “sushi-like” lipid-nanodiscs (right panel). As reported in our publications [79, 87, 88], the ability of a synthetic polymer to solubilize lipid aggregates is characterized by static light scattering (SLS) experiments on multilamellar vesicles (MLVs), TIRF based fluorescence experiments and phosphorus-31 NMR experiments. Polymer-based nanodiscs are prepared via the self-assembly process by directly mixing an appropriate ratio of polymer and lipid(s) in a buffer. Then, the mixture is incubated and purified by size-exclusion chromatography (SEC). The purified nanodiscs are characterized by dynamic light scattering (DLS) experiments to determine the size distribution and transmission electron microscopy (TEM) images to evaluate the size homogeneity of polymer-based lipid-nanodiscs. Finally, the isotropic nanodiscs are further characterized by solution NMR spectroscopy while the anisotropic macro-nanodiscs are analyzed using a variety of solid-state NMR experiments including 31P, 14N, 2D 1H/1H RFDR, SLF and 2D 1H/13C HETCOR. Our experience in the preparation of polymer-based nanodiscs suggests the use of freeze-thaw cycles for some of the lipids including POPC, POPG and DSPC as needed. In addition, sample preparation procedures for a successful reconstitution of a membrane protein need to be adapted based on the physicochemical properties of the protein under investigation. For a given membrane protein, magnetic-alignment of macro-nanodiscs and successful implementation of solid-state NMR experiments require optimization of various parameters including the lipid:polymer ratio, concentration of nanodiscs, amount of paramagnetic (for example, lanthanide) salt and temperature.
2.1. Expanding the range of sizes nanodiscs by using SMA-EA.
The first functionalized SMA based polymer we developed was styrene maleic anhydride – ethanol amine (SMA-EA) [79] using the starting material poly(styrene-co-maleic anhydride) cumene terminated with an approximately 1.3:1 styrene:maleic anhydride molar ratio with a number average molecular weight of about ~1.6 kDa. SMA-EA was synthesized by modifying the starting SMAnh via a nucleophilic ring opening reaction using ethanol amine (Figure 1). The resulting polymer was shown to form nanodiscs with a large range of sizes (10–60 nm diameter) by varying the lipid to polymer ratio. Small SMA-EA nanodiscs, with lipid:polymer (w/w) ~ 2, (<20 nm) were shown to be suitable for solution NMR experiments, whereas the larger sized nanodiscs (macro-nanodiscs) (lipid:polymer 1:1 w/w) aligned in the presence of an external magnetic field enabling structural studies of membrane proteins using solid-state NMR spectroscopy [80–82]. The ability of nanodiscs to be used in the study of membrane proteins using solution as well as solid state NMR were shown using Cyt b5 as an example. Uniformly-15N-labeled Cytochrome-b5 reconstituted in small nanodiscs (10 nm) exhibited well dispersed peaks in a 2D TROSY-HSQC (transverse relaxation optimized spectroscopy-heteronuclear single quantum correlation) spectrum suggesting that protein is well folded. Cyt b5 reconstituted in macro-nanodiscs were used in a 2D PISEMA (polarization inversion and spin exchange at magic angle) experiment [81, 83–86]. The PISEMA spectrum revealed a characteristic wheel-like pattern of resonances showing the tilt of the helical transmembrane domain with respect to the lipid bilayer normal. The SMA-EA polymer was also shown to have an increased stability towards divalent metal ions (up to 21 mM for Ca2+ and 30 mM for Mg2+), and an increased tolerance towards low pH (from pH 4.5 to pH 3.3 based on the lipid:polymer w/w ratio)) as compared to SMALP (pH ~ 6.3). While we demonstrated that SMA-EA polymer nanodiscs could be formed in a wide variety of sizes, the presence of carboxylic groups as the hydrophilic component still limited the application of SMA-EA nanodiscs.
2.2. Acidic compatible polymer nanodiscs using SMA-ED and SMAd-A.
Because the SMAnh anhydride group is easily functionalized by amines, we hypothesized that a substitution of the carboxylic acids with other hydrophilic groups would increase the stability of the polymer nanodiscs towards low pH. To test this hypothesis, we synthesized two different SMA derivatives, styrene maleic acid – ethylene diamine (SMA-ED) and styrene maleimide – amine (SMAd-A) [87]. First, we synthesized SMA-ED, a zwitterionic analog of SMA, with the idea that the polymer would always be charged due to the negatively charged carboxylates at high pH and the positive charged amino groups at low pH. This polymer showed stability for a wide range of pH values (3.5 < pH < 8.5) during the presence of only one kind of charged functional group. One interesting result was that at near neutral environment (pH ~ 6 ± 1) SMA-ED was not stable and precipitated from solution. We interpreted this observation as being a result of the zwitterionic state of SMA-ED forming aggregates with itself due to the presence of attractive charge-charge interactions. SMAd-A was synthesized to show a monofunctional form of SMA that is stable at low pH. The SMAd-A polymer was formed from a dehydration reaction of a Boc-protected SMA-ED to form the maleimide, followed by a Boc deprotection. This positively charged SMAd-A demonstrated the expected characteristic stability at acidic conditions (pH ~ 3.5) while it precipitated at basic conditions (pH > 6) due to the loss of charge on the amino functional groups. SMA-ED and SMAd-A both showed a remarkable stability towards the presence of divalent metal ions at acidic pH. These results demonstrated that the replacement of the metal chelating carboxylates with non-chelating amino groups allows for the formation of nanodiscs even in the presence of divalent metal ions and at lower pH. Both SMA-ED and SMAd-A based lipid nanodiscs have also been demonstrated to stabilize a medically important polyphenolic compound, curcumin, showing a potential application in the field of drug delivery [87].
2.3. Robust pH resistant nanodiscs using SMA-QA
Both SMA-ED and SMAd-A still had limitations due to the presence of pH dependent charged hydrophilic groups, that restricted their use to specific pH ranges. With the goal to form polymer nanodiscs that are stable under all biologically relevant pH and metal ion concentrations, we synthesized styrene maleimide – quaternary ammonium (SMA-QA) polymer [88]. We selected a quaternary ammonium as the charged hydrophilic group due to its pH independent charge and nonchelating properties. SMA-QA was synthesized using a similar approach previously seen in our SMAd-A polymer synthesis. The functionalization was performed by reacting SMA (~1.6 kDa) with aminoethyltrimethylammonium chloride hydrochloride followed by a dehydration reaction forming the maleimide. SMA-QA was shown to form monodispersed nanodiscs of a variety of sizes (10–30 nm) by varying the lipid to polymer ratio (1:1.5 to 1:0.25 w/w). These nanodiscs were shown to be monodispersed in size as characterized by dynamic light scattering (DLS), transmission electron microscopy (TEM), and size exclusion chromatography (SEC). The smaller nanodiscs (~10 nm) were shown to be isotropic in solution, enabling solution NMR studies on membrane proteins, whereas the macro-nanodiscs (>20 nm) exhibited spontaneous alignment in the magnetic field and are suitable for solid-state NMR studies. The remarkable feature of SMA-QA nanodiscs is that the nanodiscs are stable in a wide range of pH values (2.5 < pH <10) and in the presence of divalent metal ions (up to 200 mM of Ca2+ or Mg2+). Because of these unique properties, SMA-QA nanodiscs are a robust membrane mimetic tool that offers significant advantages over all currently reported nanodisc systems, and therefore we foresee a significant expansion in the applicability of nanodisc technology.
Recent studies have demonstrated the many advantages of SMA based polymers over peptidic based nanodiscs, which includes studies on membrane proteins.[71, 89–94] But, the functional modifications described here would drastically increase the applications of these polymer based nanodiscs in studying membrane proteins using a wide variety of biophysical techniques including solution and solid-state NMR spectroscopy. Most of the polymer nanodiscs studies on membrane proteins reported in the literature is limited to neutral pH, whereas SMA-QA polymer can be used to study the pH dependent membrane insertion or function of a membrane protein even at low pH [61, 95]. we also would like to mention that a strong UV absorption and the interactions with aromatic styrene group of SMA based polymers SMA based polymers pose limitations for the applications of some of the biophysical techniques. For example, the SMA polymer has been shown to have strong interactions with Thioflavin T (ThT) dye that is commonly used in monitoring the kinetics of amyloid aggregation. To overcome these limitations of polymer based nanodiscs, recent developments have focused on the production of styrene-free polymers, which include Diisobutylene/Maleic Acid Copolymer[69] and Polymethacrylate Copolymers.[96]
3. Conclusions and future directions
In conclusion functionalization of a low molecular weight SMA polymer allowed for the tuning and enhancement of these polymers to differing pH and metal ion presence while also allowing for a greater control over size. Using size control, the applications of polymer nanodiscs in the field of NMR spectroscopy, specifically for structural studies using solid-state NMR spectroscopy, is achievable. We expect that macro-nanodiscs will prove to be a valuable tool in the study of functional reconstitution and structural investigation of large-size membrane proteins and membrane protein assemblies like channels and complexes. These macro-nanodiscs will allow the use of sophisticated biophysical techniques including cryo-electron microscopy, SAXS and SANS. Overall the ability to use a straightforward chemical functionalization to modify the functional groups of SMA polymers enable us to engineer new polymers to overcome the current limitations seen in polymer nanodiscs technology. We have also demonstrated the use of styrene-free polymers to form lipid-nanodiscs which have been shown to be potentially useful in the investigation of amyloid aggregation and for studies using styrene-sensitive biophysical experiments such as circular dichroism (CD) and thioflavin-T (ThT) based fluorescence [96]. This allows us to greatly expand the use of polymer nanodiscs for biophysical structural and functional studies on a variety of membrane proteins, membrane-bound protein-protein complexes, and domains of cell membranes. It is also worthwhile to explore the feasibilities of potential biomedical applications of polymer based nanodiscs.
We believe that further optimization of the polymers would be useful to continue to expand the applications of polymer nanodiscs. Some of the polymer aspects worth investigating include the following. First, the optimization of the styrene and maleic acid group alternation in the chain (perfect alternating versus statistically random) may provide additional insights into protein structural biology using nanodiscs. Second, the design of a variety of hydrophobic groups to accommodate the variation in the hydrophobic thickness of the lipid bilayer while eliminating the styrene moiety due to its potential interaction with proteins. Third, it is also important to further investigate the lamellar nature of lipids in SMA based nanodiscs using calorimetry and solid-state NMR based experiments, and to systematically investigate the effect of the hydrophobic groups of the polymers on the structure and function of the reconstituted protein(s) and on the physicochemical properties of lipids. Finally, further studies to optimize the reconstitution of a variety of membrane composition, asymmetric lipid bilayer, and raft-like domains would further broaden the scope of polymer-based nanodiscs.
Figure 2. pH stability of polymer nanodiscs.
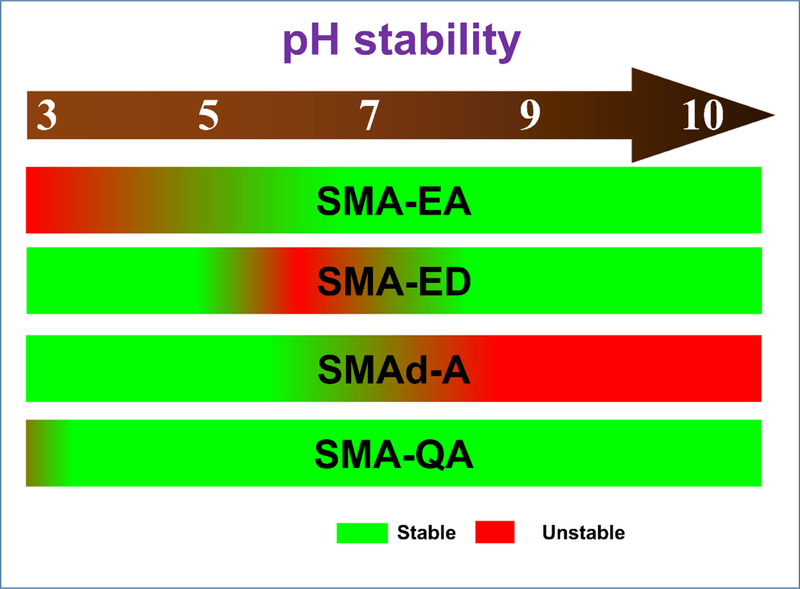
A schematic representation of the stability of various polymer nanodiscs under different pH based on our experimental characterization as reported elsewhere.[79, 87, 88]
Acknowledgements.
This study was supported by funds from NIH (GM084018 and AG048934 to A.R.). Funds from NIH to upgrade the 400 MHz solid-state NMR spectrometer are also acknowledged. A.R. thanks Dr. Antoinette Killian for fruitful discussion on SMALPs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ye L, Neale C, Sljoka A, Lyda B, Pichugin D, Tsuchimura N, Larda ST, Pomes R, Garcia AE, P Ernst O, Sunahara RK, Prosser RS, Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations. Nat Commun 9 (2018) 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Prosser RS, Ye L, Pandey A, Orazietti A, Activation processes in ligand-activated G protein-coupled receptors: A case study of the adenosine A2A receptor. Bioessays 39 (2017) 1700072. [DOI] [PubMed] [Google Scholar]
- [3].Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS, Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533 (2016) 265–268. [DOI] [PubMed] [Google Scholar]
- [4].Cournia Z, Allen TW, Andricioaei I, Antonny B, Baum D, Brannigan G, Buchete NV, Deckman JT, Delemotte L, Del Val C, Friedman R, Gkeka P, Hege HC, Henin J, Kasimova MA, Kolocouris A, Klein ML, Khalid S, Lemieux MJ, Lindow N, Roy M, Selent J, Tarek M, Tofoleanu F, S Vanni, Urban S, Wales DJ, Smith JC, Bondar AN, Membrane Protein Structure, Function, and Dynamics: a Perspective from Experiments and Theory. J Membr Biol 248 (2015) 611–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fry MY, Clemons WM Jr., Complexity in targeting membrane proteins. Science 359 (2018) 390–391. [DOI] [PubMed] [Google Scholar]
- [6].Wallin E, von Heijne G, Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 7 (1998) 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Overington JP, Al-Lazikani B, Hopkins AL, How many drug targets are there? Nat Rev Drug Discov 5 (2006) 993–996. [DOI] [PubMed] [Google Scholar]
- [8].Rawson S, Davies S, Lippiat JD, Muench SP, The changing landscape of membrane protein structural biology through developments in electron microscopy. Mol Membr Biol 33 (2016) 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Parmar MJ, Lousa Cde M, Muench SP, Goldman A, Postis VL, Artificial membranes for membrane protein purification, functionality and structure studies. Biochem Soc Trans 44 (2016) 877–882. [DOI] [PubMed] [Google Scholar]
- [10].Schreiber SL, Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287 (2000) 1964–1969. [DOI] [PubMed] [Google Scholar]
- [11].Anandan A, Vrielink A, in The Next Generation in Membrane Protein Structure Determination (Ed.: Moraes I), Springer International Publishing, Cham, 2016, pp. 13–28. [Google Scholar]
- [12].Chipot C, Dehez F, Schnell JR, Zitzmann N, Pebay-Peyroula E, Catoire LJ, Miroux B, Kunji ERS, Veglia G, Cross TA, Schanda P, Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem Rev 118 (2018) 3559–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Arnold T, Linke D, The use of detergents to purify membrane proteins. Curr Protoc Protein Sci. Chapter 4 (2008) Unit 4 8 1–4 8 30. [DOI] [PubMed] [Google Scholar]
- [14].Garavito RM, Ferguson-Miller S, Detergents as tools in membrane biochemistry. J Biol Chem 276 (2001) 32403–32406. [DOI] [PubMed] [Google Scholar]
- [15].Helenius A, Simons K, Solubilization of membranes by detergents. Biochim Biophys Acta 415 (1975) 29–79. [DOI] [PubMed] [Google Scholar]
- [16].Tanford C, Reynolds JA, Characterization of membrane proteins in detergent solutions. Biochim Biophys Acta 457 (1976) 133–170. [DOI] [PubMed] [Google Scholar]
- [17].Bordier C, Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256 (1981) 1604–1607. [PubMed] [Google Scholar]
- [18].Seddon AM, Curnow P, Booth PJ, Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta 1666 (2004) 105–117. [DOI] [PubMed] [Google Scholar]
- [19].Linke D, in Methods in Enzymology, Vol. 463 (Eds.: Burgess RR, Deutscher MP), Academic Press, 2009, pp. 603–617.19892194 [Google Scholar]
- [20].Rigaud J-L, Lévy D, in Methods in Enzymology, Vol. 372, Academic Press, 2003, pp. 65–86. [DOI] [PubMed] [Google Scholar]
- [21].Rigaud JL, Paternostre MT, Bluzat A, Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry 27 (1988) 2677–2688. [DOI] [PubMed] [Google Scholar]
- [22].Durr UH, Gildenberg M, Ramamoorthy A, The magic of bicelles lights up membrane protein structure. Chem Rev 112 (2012) 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sanders CR, Prosser RS, Bicelles: a model membrane system for all seasons? Structure 6 (1998) 1227–1234. [DOI] [PubMed] [Google Scholar]
- [24].Sanders CR, Landis GC, Reconstitution of Membrane Proteins into Lipid-Rich Bilayered Mixed Micelles for NMR Studies. Biochemistry 34 (1995) 4030–4040. [DOI] [PubMed] [Google Scholar]
- [25].Soong R, Xu J, Ramamoorthy A, in Nuclear Magnetic Resonance Spectroscopy of Liquid Crystals, pp. 117–128.
- [26].Beaugrand M, Arnold AA, Henin J, Warschawski DE, Williamson PT, Marcotte I, Lipid concentration and molar ratio boundaries for the use of isotropic bicelles. Langmuir 30 (2014) 6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sanders CR, Schwonek JP, Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry 31 (1992) 8898–8905. [DOI] [PubMed] [Google Scholar]
- [28].Sanders CR, Hare BJ, Howard KP, Prestegard JH, Magnetically-Oriented Phospholipid Micelles as a Tool for the Study of Membrane-Associated Molecules. Prog Nucl Mag Res Sp 26 (1994) 421–444. [Google Scholar]
- [29].Cardon TB, Dave PC, Lorigan GA, Magnetically Aligned Phospholipid Bilayers with LargeqRatios Stabilize Magnetic Alignment with High Order in the Gel and LαPhases. Langmuir 21 (2005) 4291–4298. [DOI] [PubMed] [Google Scholar]
- [30].Yamamoto K, Soong R, Ramamoorthy A, Comprehensive analysis of lipid dynamics variation with lipid composition and hydration of bicelles using nuclear magnetic resonance (NMR) spectroscopy. Langmuir 25 (2009) 7010–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Denisov IG, Sligar SG, Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol 23 (2016) 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bayburt TH, Grinkova YV, Sligar SG, Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters 2 (2002) 853–856. [Google Scholar]
- [33].Bayburt TH, Carlson JW, Sligar SG, Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J Struct Biol 123 (1998) 37–44. [DOI] [PubMed] [Google Scholar]
- [34].Nath A, Atkins WM, Sligar SG, Applications of Phospholipid Bilayer Nanodiscs in the Study of Membranes and Membrane Proteins. Biochemistry 46 (2007) 2059–2069. [DOI] [PubMed] [Google Scholar]
- [35].Denisov IG, Sligar SG, Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev 117 (2017) 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hagn F, Etzkorn M, Raschle T, Wagner G, Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. J. Am. Chem. Soc 135 (2013) 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, Wagner G, Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 14 (2017) 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG, in Methods in Enzymology, Vol. 464 (Ed.: Düzgünes N), Academic Press, 2009, pp. 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hagn F, Nasr ML, Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat Protoc 13 (2018) 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rodriguez Camargo DC, Korshavn KJ, Jussupow A, Raltchev K, Goricanec D, Fleisch M, Sarkar R, Xue K, Aichler M, Mettenleiter G, Walch AK, Camilloni C, Hagn F, Reif B, Ramamoorthy A, Stabilization and structural analysis of a membrane-associated hIAPP aggregation intermediate. Elife 6 (2017) e31226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nath A, Miranker AD, Rhoades E, A membrane-bound antiparallel dimer of rat islet amyloid polypeptide. Angew Chem Int Ed Engl 50 (2011) 10859–10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A, Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies Using NMR Spectroscopy. Angew. Chem. Int. Ed 128 (2016) 4497–4499. [DOI] [PubMed] [Google Scholar]
- [43].Ravula T, Barnaba C, Mahajan M, Anantharamaiah GM, Im SC, Waskell L, Ramamoorthy A, Membrane environment drives cytochrome P450’s spin transition and its interaction with cytochrome b5. Chem Commun (Camb) 53 (2017) 12798–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM, Ramamoorthy A, Real-Time Monitoring of Lipid Exchange via Fusion of Peptide Based Lipid-Nanodiscs. Chem Mater 30 (2018) 3204–3207. [Google Scholar]
- [45].Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im SC, Anantharamaiah GM, Waskell L, Ramamoorthy A, Cytochrome-P450-Induced Ordering of Microsomal Membranes Modulates Affinity for Drugs. Angew Chem Int Ed Engl 57 (2018) 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Barnaba C, Ravula T, Medina-Meza IG, Im SC, Anantharamaiah GM, Waskell L, Ramamoorthy A, Lipid-exchange in nanodiscs discloses membrane boundaries of cytochrome-P450 reductase. Chem Commun (Camb) 54 (2018) 6336–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Prade E, Mahajan M, Im SC, Zhang M, Gentry KA, Anantharamaiah GM, Waskell L, Ramamoorthy A, A Minimal Functional Complex of Cytochrome P450 and FBD of Cytochrome P450 Reductase in Nanodiscs. Angew Chem Int Ed Engl 57 (2018) 8458–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ, Nanodiscs versus Macrodiscs for NMR of Membrane Proteins. Biochemistry 50 (2011) 8983–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ, Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater 16 (2017) 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kuai R, Li D, Chen YE, Moon JJ, Schwendeman A, High-Density Lipoproteins: Nature’s Multifunctional Nanoparticles. ACS Nano 10 (2016) 3015–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Scheidelaar S, Koorengevel MC, van Walree CA, Dominguez JJ, Dörr JM, Killian AJ, Effect of Polymer Composition and pH on Membrane Solubilization by Styrene-Maleic Acid Copolymers. Biophys. J 111 (2016) 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA, Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. U.S.A 111 (2014) 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Grobner G, Watts A, Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew. Chem. Int. Ed. Engl 51 (2012) 4653–4657. [DOI] [PubMed] [Google Scholar]
- [54].Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR, A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc 11 (2016) 1149–1162. [DOI] [PubMed] [Google Scholar]
- [55].Knowles TJ, Finka R, Smith C, Lin Y-P, Dafforn T, Overduin M, Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc 131 (2009) 7484–7485. [DOI] [PubMed] [Google Scholar]
- [56].Rajesh S, Knowles T, Overduin M, Production of membrane proteins without cells or detergents. New Biotechnol 28 (2011) 250–254. [DOI] [PubMed] [Google Scholar]
- [57].Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas OR, Overduin M, Dafforn TR, Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem Soc Trans 39 (2011) 813–818. [DOI] [PubMed] [Google Scholar]
- [58].Web link for polymer nanodisc: http://www.smalp.net/.
- [59].Stroud Z, Hall SCL, Dafforn TR, Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights. Methods (2018). [DOI] [PubMed]
- [60].Xie Z, Schendel S, Matsuyama S, Reed JC, Acidic pH Promotes Dimerization of Bcl-2 Family Proteins. Biochemistry 37 (1998) 6410–6418. [DOI] [PubMed] [Google Scholar]
- [61].O’Keefe DO, Cabiaux V, Choe S, Eisenberg D, Collier RJ, pH-dependent insertion of proteins into membranes: B-chain mutation of diphtheria toxin that inhibits membrane translocation, Glu-349----Lys. Proc. Natl. Acad. Sci. U.S.A 89 (1992) 6202–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu Y, Bhate MP, McDermott AE, Transmembrane allosteric energetics characterization for strong coupling between proton and potassium ion binding in the KcsA channel. Proc Natl Acad Sci U S A 114 (2017) 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Trivedi BC, Culbertson BM, Maleic Anhydride, Springer S, Springer; (1982). [Google Scholar]
- [64].Swainsbury DJK, Scheidelaar S, van Grondelle R, Killian JA, Jones MR, Bacterial Reaction Centers Purified with Styrene Maleic Acid Copolymer Retain Native Membrane Functional Properties and Display Enhanced Stability. Angew. Chem. Int. Ed. Engl 53 (2014) 11803–11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tanaka M, Hosotani A, Tachibana Y, Nakano M, Iwasaki K, Kawakami T, Mukai T, Preparation and Characterization of Reconstituted Lipid–Synthetic Polymer Discoidal Particles. Langmuir 31 (2015) 12719–12726. [DOI] [PubMed] [Google Scholar]
- [66].Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schäfer M, van Walree CA, Killian JA, The styrene–maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J 45 (2016) 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Morrison KA, Akram A, Mathews A, Khan ZA, Patel JH, Zhou C, Hardy DJ, Moore-Kelly C, Patel R, Odiba V, Knowles TJ, Javed M.-u.-H., Chmel NP, Dafforn TR, Rothnie AJ, Membrane protein extraction and purification using styrene–maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem. J 473 (2016) 4349–4360. [DOI] [PubMed] [Google Scholar]
- [68].Hu Z, Ho JCS, Nallani M, Synthetic (polymer) biology (membrane): functionalization of polymer scaffolds for membrane proteins. Curr. Opin. Biotechnol 46 (2017) 51–56. [DOI] [PubMed] [Google Scholar]
- [69].Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S, Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew Chem Int Ed Engl 56 (2017) 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB, Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557 (2018) 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, Silvestro D, Hatzakis NS, Wenk MR, Dafforn TR, Olsen CE, Motawia MS, Hamberger B, Moller BL, Bassard JE, Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 354 (2016) 890–893. [DOI] [PubMed] [Google Scholar]
- [72].Morrison KA, Akram A, Mathews A, Khan ZA, Patel JH, Zhou C, Hardy DJ, Moore-Kelly C, Patel R, Odiba V, Knowles TJ, Javed MU, Chmel NP, Dafforn TR, Rothnie AJ, Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem J 473 (2016) 4349–4360. [DOI] [PubMed] [Google Scholar]
- [73].Grethen A, Oluwole AO, Danielczak B, Vargas C, Keller S, Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Sci Rep 7 (2017) 11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cuevas Arenas R, Klingler J, Vargas C, Keller S, Influence of lipid bilayer properties on nanodisc formation mediated by styrene/maleic acid copolymers. Nanoscale 8 (2016) 15016–15026. [DOI] [PubMed] [Google Scholar]
- [75].Hall SCL, Tognoloni C, Price GJ, Klumperman B, Edler KJ, Dafforn TR, Arnold T, Influence of Poly(styrene- co-maleic acid) Copolymer Structure on the Properties and Self-Assembly of SMALP Nanodiscs. Biomacromolecules 19 (2018) 761–772. [DOI] [PubMed] [Google Scholar]
- [76].Zhang R, Sahu ID, Bali AP, Dabney-Smith C, Lorigan GA, Characterization of the structure of lipodisq nanoparticles in the presence of KCNE1 by dynamic light scattering and transmission electron microscopy. Chem Phys Lipids 203 (2017) 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Martinez D, Decossas M, Kowal J, Frey L, Stahlberg H, Dufourc EJ, Riek R, Habenstein B, Bibow S, Loquet A, Lipid Internal Dynamics Probed in Nanodiscs. Chemphyschem 18 (2017) 2651–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Puthenveetil R, Nguyen K, Vinogradova O, Nanodiscs and Solution NMR: preparation, application and challenges. Nanotechnol Rev 6 (2017) 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A, Bioinspired, Size-Tunable Self-Assembly of Polymer-Lipid Bilayer Nanodiscs. Angew Chem Int Ed Engl 56 (2017) 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [80].Yamamoto K, Gildenberg M, Ahuja S, Im SC, Pearcy P, Waskell L, Ramamoorthy A, Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles. Sci Rep 3 (2013) 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ramamoorthy A, Wei Y, Lee D-K, in Annual Reports on NMR Spectroscopy, Vol. 52, Academic Press, 2004, pp. 1–52. [Google Scholar]
- [82].Radoicic J, Park SH, Opella SJ, Macrodiscs Comprising SMALPs for Oriented Sample Solid-State NMR Spectroscopy of Membrane Proteins. Biophys J 115 (2018) 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gayen A, Banigan JR, Traaseth NJ, Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew Chem Int Ed Engl 52 (2013) 10321–10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang S, Gopinath T, Veglia G, Application of paramagnetic relaxation enhancements to accelerate the acquisition of 2D and 3D solid-state NMR spectra of oriented membrane proteins. Methods 138–139 (2018) 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Salnikov ES, Anantharamaiah GM, Bechinger B, Supramolecular Organization of Apolipoprotein-A-I-Derived Peptides within Disc-like Arrangements. Biophys J 115 (2018) 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Salnikov ES, Sarrouj H, Reiter C, Aisenbrey C, Purea A, Aussenac F, Ouari O, Tordo P, Fedotenko I, Engelke F, Bechinger B, Solid-State NMR/Dynamic Nuclear Polarization of Polypeptides in Planar Supported Lipid Bilayers. J Phys Chem B 119 (2015) 14574–14583. [DOI] [PubMed] [Google Scholar]
- [87].Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A, pH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs. Langmuir 33 (2017) 10655–10662. [DOI] [PubMed] [Google Scholar]
- [88].Ravula T, Hardin NZ, Ramadugu SK, Cox SJ, Ramamoorthy A, Formation of pH-Resistant Monodispersed Polymer-Lipid Nanodiscs. Angew Chem Int Ed Engl 57 (2018) 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [89].Logez C, Damian M, Legros C, Dupre C, Guery M, Mary S, Wagner R, M’Kadmi C, Nosjean O, Fould B, Marie J, Fehrentz JA, Martinez J, Ferry G, Boutin JA, Baneres JL, Detergent-free Isolation of Functional G Protein-Coupled Receptors into Nanometric Lipid Particles. Biochemistry 55 (2016) 38–48. [DOI] [PubMed] [Google Scholar]
- [90].Smirnova IA, Sjostrand D, Li F, Bjorck M, Schafer J, Ostbye H, Hogbom M, von Ballmoos C, Lander GC, Adelroth P, Brzezinski P, Isolation of yeast complex IV in native lipid nanodiscs. Biochim Biophys Acta 1858 (2016) 2984–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Komar J, Alvira S, Schulze RJ, Martin R, Lycklama ANJA, Lee SC, Dafforn TR, Deckers-Hebestreit G, Berger I, Schaffitzel C, Collinson I, Membrane protein insertion and assembly by the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Biochem J 473 (2016) 3341–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bersch B, Dorr JM, Hessel A, Killian JA, Schanda P, Proton-Detected Solid-State NMR Spectroscopy of a Zinc Diffusion Facilitator Protein in Native Nanodiscs. Angew Chem Int Ed Engl 56 (2017) 2508–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].McDowall JS, Ntai I, Hake J, Whitley PR, Mason JM, Pudney CR, Brown DR, Steady-State Kinetics of alpha-Synuclein Ferrireductase Activity Identifies the Catalytically Competent Species. Biochemistry 56 (2017) 2497–2505. [DOI] [PubMed] [Google Scholar]
- [94].Ravula T, Hardin NZ, Bai J, Im SC, Waskell L, Ramamoorthy A, Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs. Chem Commun (Camb) 54 (2018) 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wolf J, Aisenbrey C, Harmouche N, Raya J, Bertani P, Voievoda N, Suss R, Bechinger B, pH-Dependent Membrane Interactions of the Histidine-Rich Cell-Penetrating Peptide LAH4-L1. Biophys J 113 (2017) 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi JI, Ramamoorthy A, Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers. J Am Chem Soc 139 (2017) 18657–18663. [DOI] [PMC free article] [PubMed] [Google Scholar]


