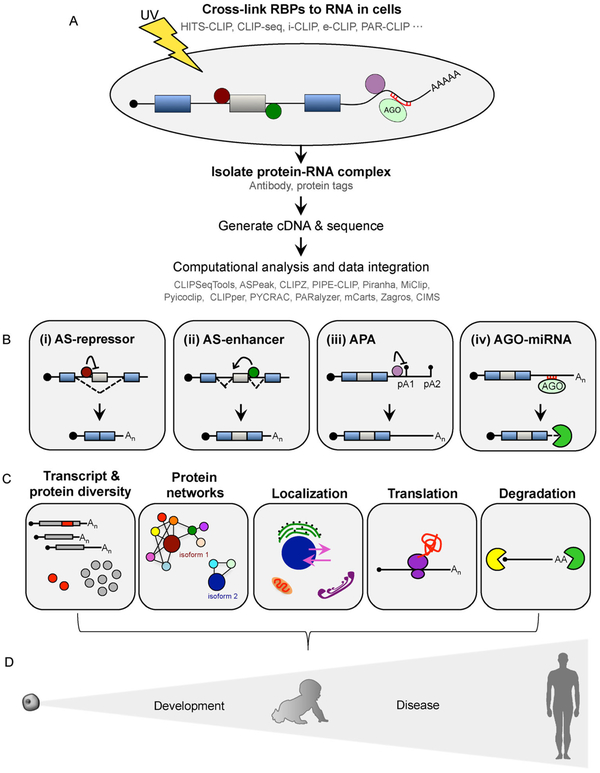
Figure 1. Integration of CLIP methods with computational analysis to explore protein-RNA interactions and their biological impact.
(A) Simplified schematic illustrating the CLIP workflow to generate transcriptome-side maps of RBP-RNA interactions occurring in vivo. (B) Integration of next generation sequencing with CLIP data can be used to identify RBPs that function as (i) repressors of alternative splicing (AS), (ii) enhancers of alternative splicing, or (iii) regulators of alternative polyadenylation (APA). CLIP can also be performed on AGO proteins to map miRNA binding sites that downregulate target mRNAs (iv). (C) Application of CLIP methods to define RBP regulatory networks from position-dependent and combinatorial functions. CLIP profiles, in conjunction with high-throughput sequencing, can characterize transcript/protein diversity in specific cellular contexts, protein networks, localization patterns, translational fates of mRNAs, and substrates of mRNA decay. (D) Identification of complex protein-RNA interaction networks that regulate gene expression is critical to understanding human development and disease.