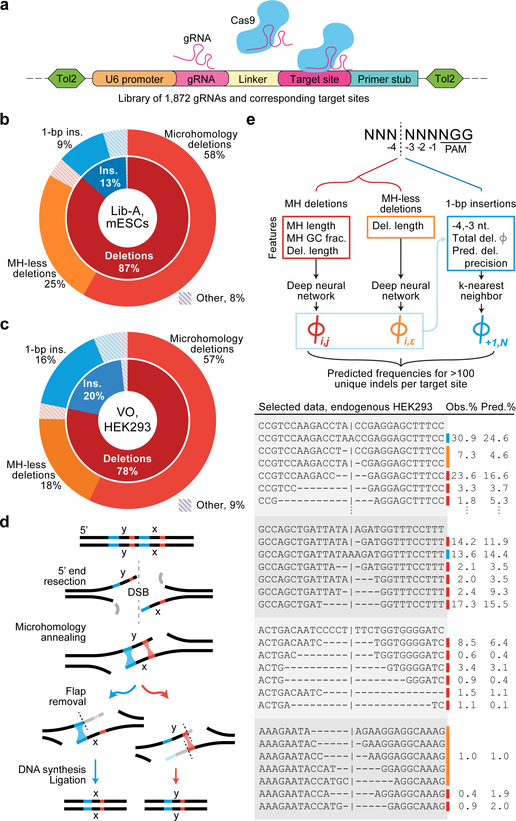
Fig. 1 |. High-throughput assaying of Cas9-mediated DNA repair products supports the design of the inDelphi model.
a, A high-throughput genome-integrated library for assaying Cas9 editing products. b, Categories of editing products at 1,996 Lib-A target sites in mouse embryonic stem cells (mESCs). c, Categories of editing products in 89 VO endogenous target sites in HEK293 cells. d, Mechanism of microhomology-mediated end-joining repair. e, inDelphi uses machine learning to predict the frequencies of editing products from target DNA sequence (selected outcomes depicted in table). Major editing outcomes include +1 to −60 indels.