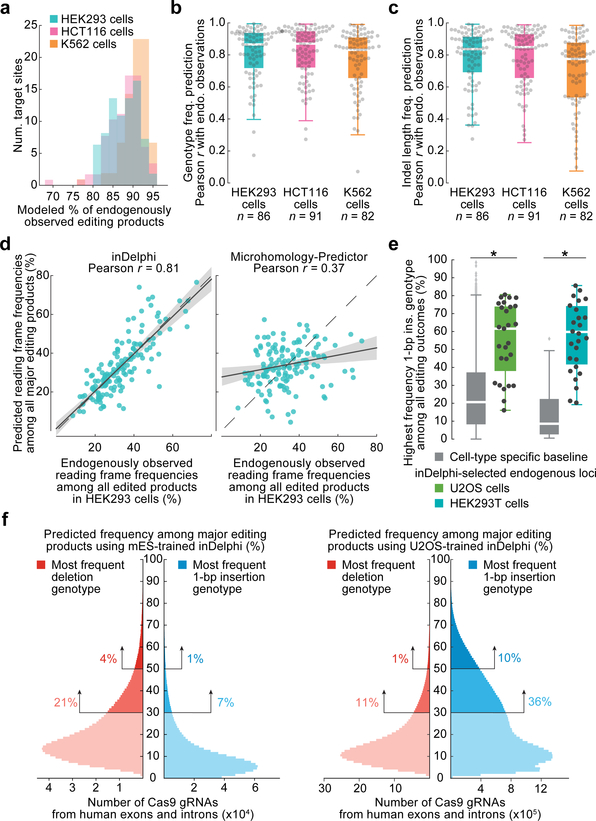
Fig. 3 |. inDelphi accurately predicts nearly all editing outcomes.
a, Fraction of endogenous editing products given predictions in HEK293 (n = 86 target sites), HCT116 (n = 91), and K562 cells (n = 82). b, c, Predictive performance on endogenously observed frequencies of genotypes (b) and indel lengths (c) in HEK293 (medians = 0.87 and 0.84), HCT116 (medians = 0.87 and 0.85), and K562 (medians = 0.83 and 0.79) cells. The box denotes the 25th, 50th, and 75th percentiles, and whiskers show 1.5 times the interquartile range. d, Comparison of predictions from two methods to observed frame frequencies (n = 86 target sites, HEK293 cells), regression estimate ± 95% C.I. e, 1-bp insertion frequencies among edited outcomes in U2OS and HEK293T cells (n = 27 and 26 observations, baseline n = 1,958 and 89 target sites, P = 4.2×10−8 and 8.1×10−12 respectively), two-sided Welch’s t-test. f, Smoothed predicted distribution of the highest frequency indel among major editing outcomes (+1 to −60 indels) for SpCas9 gRNAs targeting the human genome.