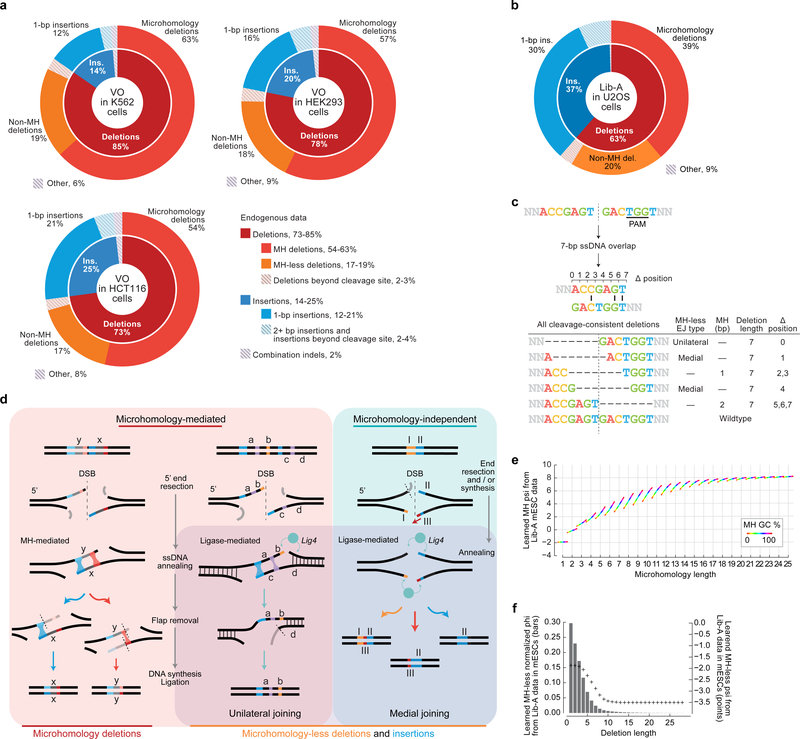
Extended Data Figure 2: Categorizing and modeling Cas9-mediated DNA repair products with manual data-analysis and automated machine learning through inDelphi.
a, b, Categories of Cas9-mediated genotypic outcomes in data from endogenous contexts at VO target sites in K562 (n = 88 target sites), HCT116 (n = 92), HEK293 (n = 89) (collectively, a) and U2OS cells (b, n = 1,958 Lib-A target sites). c, Categories and defined properties (Supplementary Methods) of all sequence alignments consistent with a Cas9-mediated 7-bp deletion. d, Hypothesized mechanisms for template-free DNA repair at Cas9-mediated DSBs based on c-NHEJ and alt-EJ/MMEJ components (Supplementary Discussion). e, Function learned for modeling MH deletions (Supplementary Methods). f, Function learned for modeling MH-independent deletions (MHless-NN) mapping deletion length to a numeric score (psi, Supplementary Methods, point plot) and with deletion length penalty normalized to sum to 1 (phi, Supplementary Methods, histogram).