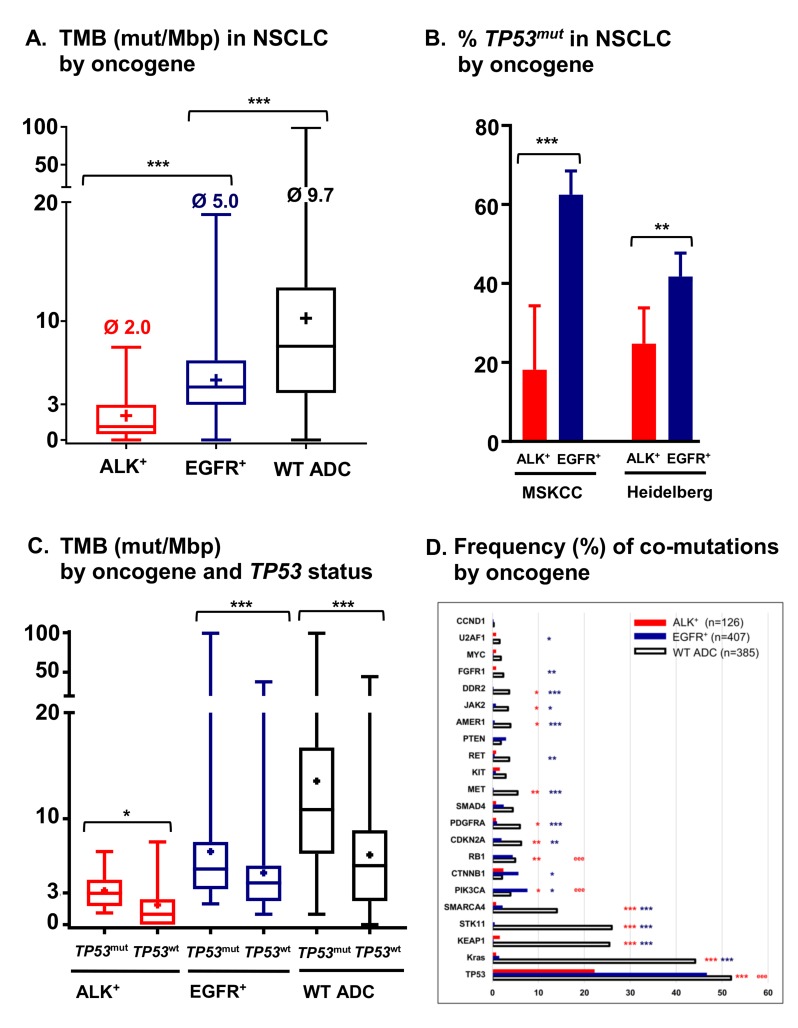
Figure 2. Tumor mutational burden, frequency of TP53 mutations and frequency of co-mutations in metastatic ALK+ and EGFR+ NSCLC.
A. Tumor mutational burden (TMB) of metastatic ALK+ (n = 33, mean 2.0 mutations[mut]/Mbp), EGFR+ (n = 232, mean 5.0 mut/Mbp) and wildtype (WT, i.e. ALK/EGFR/RET/ROS-negative, n = 557, mean 9.7 mut/Mbp) cases from the publicly available MSKCC lung adenocarcinoma (ADC) cohort (http://www.cbioportal.org) as estimated by targeted sequencing with the IMPACT341 and IMPACT411 panels [35-37]. For cases with multiple sampling time-points, only the earliest one in the disease course was analyzed, and among multiple samples at the earliest time-point, that with the highest number of mutations was chosen. Boxplots show medians, means (“+”) and range; ***p < 0.001 with the Kruskal-Wallis test followed by the Dunn’s post-hoc test. B. Frequency of TP53 mutations in metastatic ALK+ (18%, n = 33) and EGFR+ (63%, n = 232) NSCLC cases of the MSKCC cohort [35-37], as well as in untreated metastatic ALK+ (25%, n = 105) and EGFR+ (42%, n = 273) tumors sequenced for exons 4-10 of TP53 at our institution [18, 22]. Columns and error bars show percentages and 95% confidence intervals, respectively; ***p < 0.001, and **p = 0.0022 with a chi-square test. C. Tumor mutational burden (TMB) according to TP53 status for ALK+ (mean 3.2 mut/Mbp for TP53 mutated, n = 6, vs. 1.8 mut/Mbp for TP53 wildtype cases, n = 27, p = 0.039 with a Mann-Whitney test), EGFR+ (mean 5.6 mut/Mbp, n = 145, vs. 4.0 mut/Mbp, n = 87, p < 0.001) and WT cases (mean 13.6 mut/Mbp, n = 291, vs. 6.6 mut/Mbp, n = 266, p < 0.001) from the publicly available MSKCC lung adenocarcinoma cohort (http://www.cbioportal.org) [35-37]. In a bivariable linear regression analysis among ALK+ and EGFR+ patients, type of oncogene (EGFR vs. ALK, beta = 0.248, p < 0.001) and TP53 status (mutated vs. wild-type, beta = 0.256, p < 0.001) were similarly strong determinants of TMB. Boxplots show medians, means (“+”) and range. D. Frequency of co-mutations in untreated ALK+, EGFR+ and WT NSCLC patients. Analyzed were untreated ALK+ (n = 105) and EGFR+ (n = 273) patients sequenced with PCR-based DNA NGS using our custom panel of 38 genes as previously described [22], as well as the untreated ALK+ (n = 21), EGFR+ (n = 134) and WT (n = 385) patients of the MSKCC lung adenocarcinoma cohort sequenced with the MSK-IMPACT341 and MSK-IMPACT411 panels (http://www.cbioportal.org) [35-37]. Visualized are all common genes among the three panels with at least one detectable mutation. Statistical comparisons for ALK+ vs. WT, and EGFR+ vs. WT were performed with a chi-square test, and results with p < 0.05 and Benjamini-Hochberg q < 0.05 are shown in the graph in red and dark blue color, respectively: *:p < 0.05, **:p < 0.01, ***:p < 0.001. Statistical comparison for ALK+ vs. EGFR+ was performed similarly, and significant results are shown in red color: e:p < 0.05, ee:p < 0.01, eee:p < 0.001.