Figure 4.
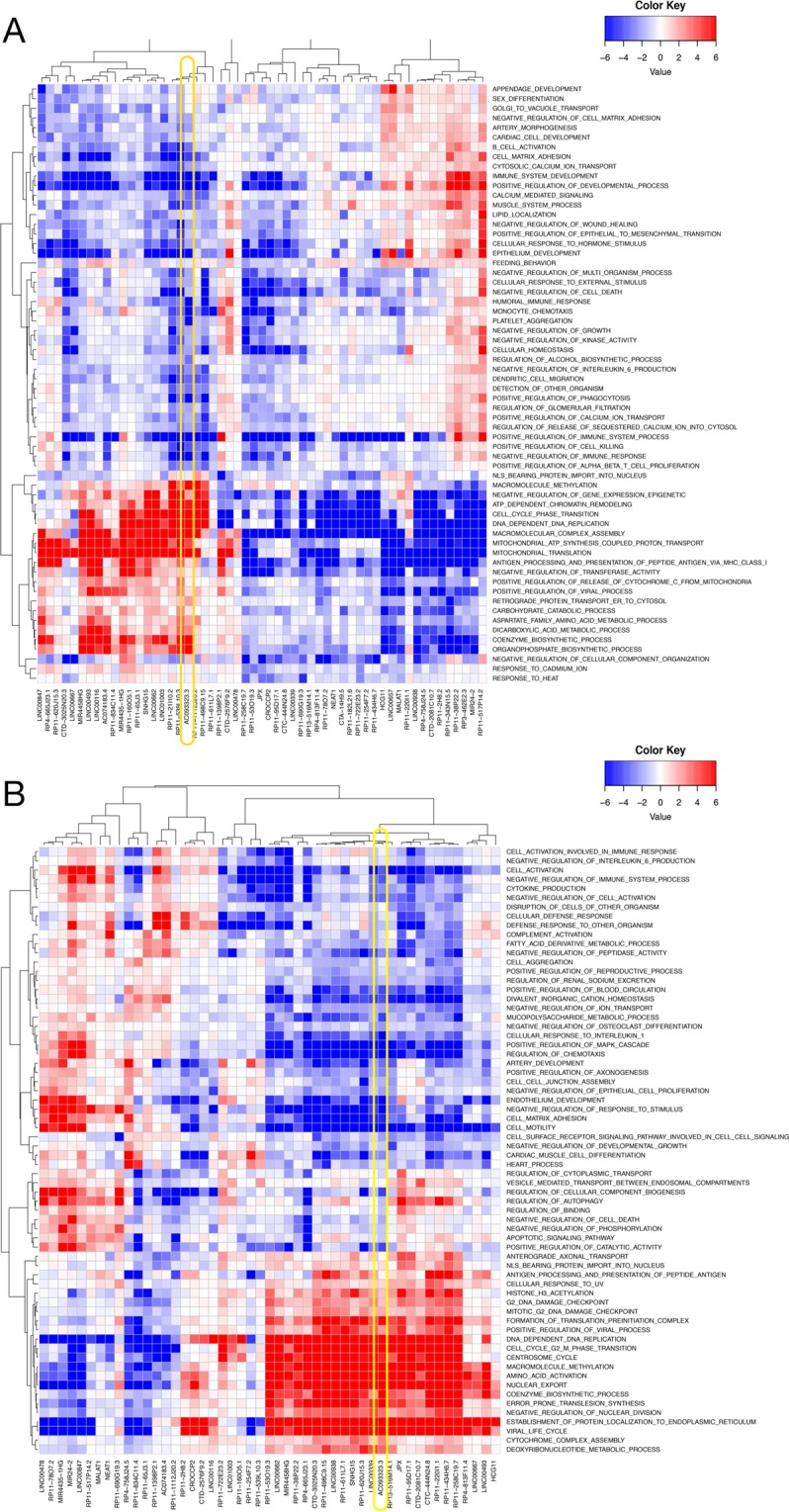
Functional prediction of HK-lincRNAs (A) HK-lincRNA function prediction using TCGA lung adenocarcinoma (LUAD)38 samples. Rows represent non-redundant Gene Ontology (GO) ‘biological process’ terms. Columns represent each HK-lincRNA. Coloured cells correspond to a significant positive (red) or negative (blue) association between an HK-lincRNA and a biological process. Associations were established based on correlation of expression level of each lincRNA with protein-coding genes. Significance was determined using Gene Set Enrichment Analysis (GSEA)39 of protein-coding genes ranked by Pearson correlation to each HK-lincRNA. (B) Functional prediction of HK-lincRNA using Encode cell lines. Rows represent non-redundant Gene Ontology (GO) biological processes terms and columns represent HK-lincRNAs. Cells are coloured as red or blue based on significant positive or negative association between HK-lincRNA and a biological process. Significant biological processes (<5% FDR) were identified for each HK- lincRNA using Gene Set Enrichment Analysis (GSEA) of protein-coding genes pre-ranked by the Pearson correlation of gene expression. Only non-redundant biological processes identified using the GOSemSim package are shown.
