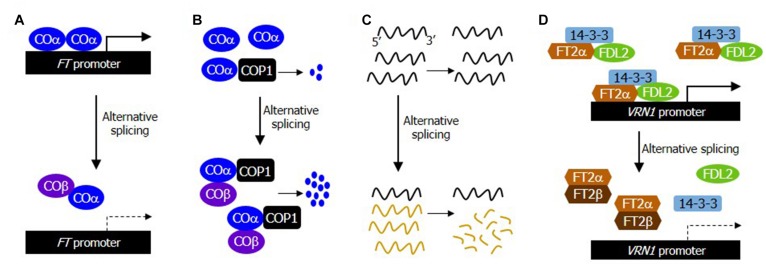
FIGURE 1.
Alternative RNA splicing regulates the timing of flowering transition through diverse regulatory mechanisms. In response to developmental and environmental cues, numerous flowering time genes undergo alternative splicing. The alternatively spliced RNA isoforms are regulated either at the RNA level through the NMD pathway or translated to multiple protein products. The alternatively spliced protein variants modulate the stability or function of different variants by forming heterodimers. They also interact differentially with their interacting partners in a competitive manner. (A) Attenuation of the DNA binding affinity of COα by forming non-DNA-binding, COα-COβ heterodimers in photoperiodic flowering. While the COα-COα homodimers are able to efficiently bind to DNA, the COα-COβ heterodimers are excluded from DNA binding. (B) Facilitation of the interaction of COα with the COP1 E3 ubiquitin ligase by COβ in photoperiodic flowering. While COα monomers are poorly targeted by COP1, COβ facilitates the interaction of COα with COP1, resulting in an elevated ubiquitin-mediated degradation of the COα proteins. (C) Differential stabilization of alternatively spliced transcripts against NMD pathway in thermosensory flowering. The alternatively spliced transcripts are degraded through the NMD pathway. (D) Attenuation of the FT2α/FDL2/14-3-3 florigenic complex formation by FT2β in aging-induced flowering time control. The alternatively spliced protein isoform FT2β inhibits the formation of the FT2α-containing florigenic complexes by forming FT2α-FT2β heterodimers in Brachypodium.