FIG 6.
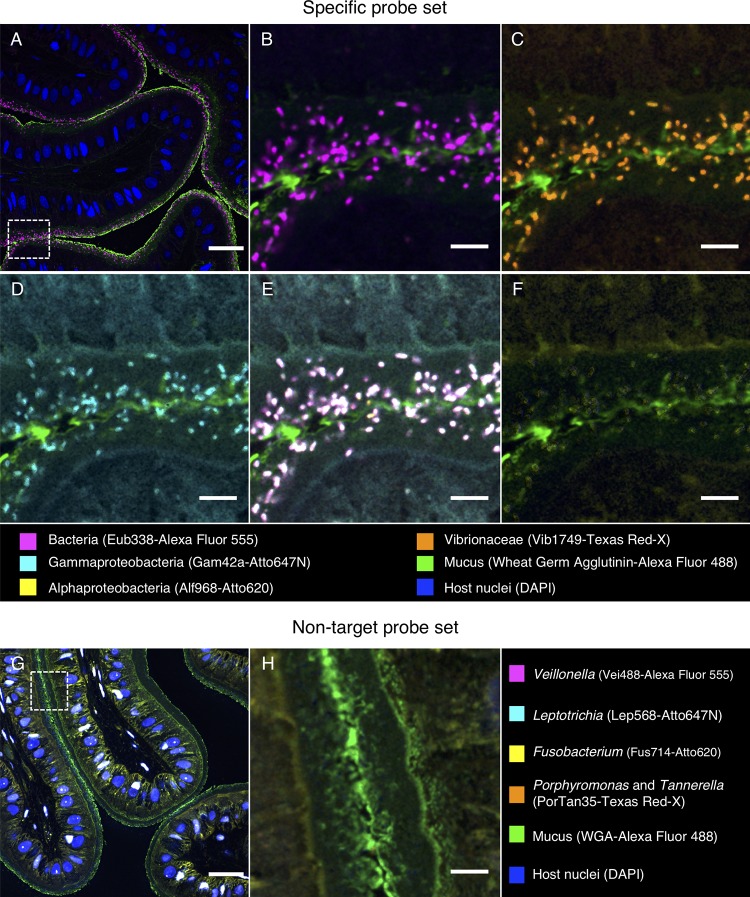
Fluorescence in situ hybridization identifies bacteria in the esophagus of S. officinalis as Vibrionaceae. A methacrylate-embedded section from a control animal was hybridized with a nested probe set containing probes for most bacteria, Gammaproteobacteria, Alphaproteobacteria, and Vibrionaceae. (A) Near-universal probe showing a bacterial distribution similar to that in Fig. 5. (B, C, and D) Enlarged images of the region marked by the dashed square in panel A showing hybridization with near-universal, Vibrionaceae, and Gammaproteobacteria probes, respectively. (E) Merged image of panels B, C, and D showing an exact match of the signal from those three probes (white). (F) Alphaproteobacteria probe showing no hybridization. (G) An independent hybridization with the nontarget probe set as a control. No signal is observed, except for nonspecific binding of probes to host cell nuclei. (H) Enlarged image of the dashed square in panel G. Scale bars = 30 μm in panels A and G and 5 μm in panels B to F and H.