Figure EV1. Topology of yeast Mpc1, Mpc2 and Mpc3.
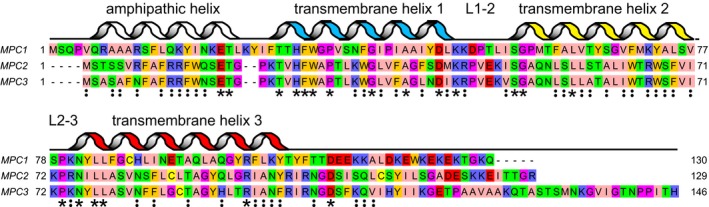
The alignment was generated by Clustal Omega and by manual curation (Sievers et al, 2011). The aligned residues are coloured by the ZAPPO colour scheme in which aliphatic, polar, aromatic, positively charged, negatively charged, Pro/Gly and Cys are coloured pink, green, orange, blue, red, magenta and yellow, respectively. The asterisks indicate identical residues and the colon conserved substitutions. Also indicated are putative transmembrane helices, loop regions and the N‐terminal amphipathic helix. The secondary structure elements were assigned based on PSIPRED (Buchan et al, 2013), MEMSAT3 (Jones et al, 1994) and conservation analysis.
