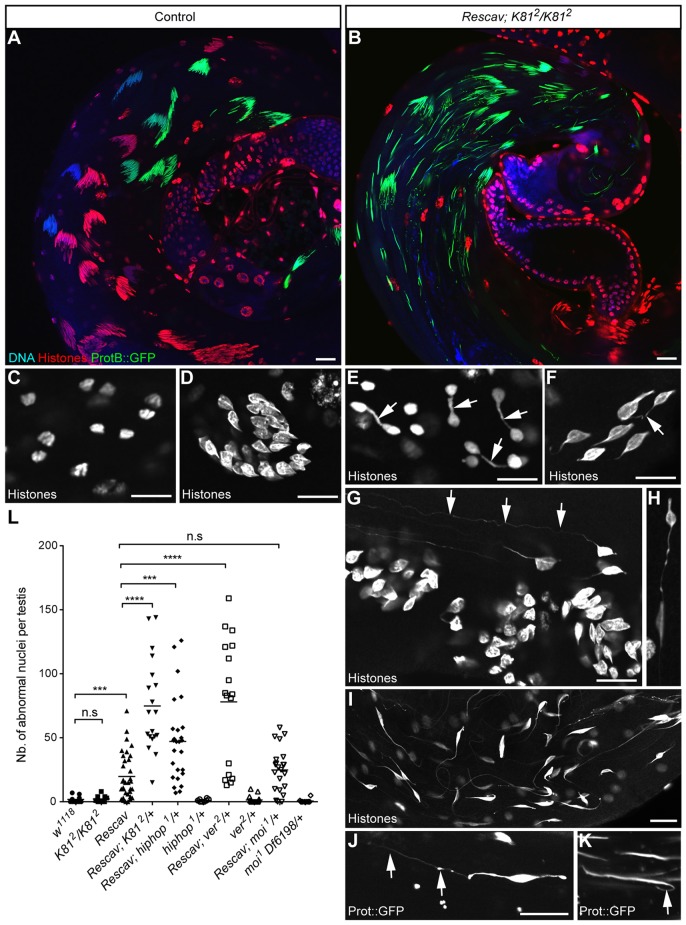
Fig. 5.
Loss of K81 in the Rescav background results in chromatin bridges at high frequency. (A,B) Representative images of a testis from a control fly bearing a protB::GFP transgene in a wild-type (A) or in a Rescav; K812/K812 (B) background stained with a pan-histone antibody (red) and DNA (blue). In Rescav; K812/K812, the cysts of spermatids are disorganized (B). (C,D) Confocal images of nuclei in testes of wild-type flies stained with an antibody against histone. (C) Anaphase of male meiosis II. (D) A group of young spermatids. (E–K) Examples of chromatin bridges observed in the Rescav; K812/K812 flies that have been stained with an antibody against histone (E–I) or expressing a protB::GFP transgene (J,K). (E) Telophase of meiosis. Arrows indicate chromosome bridges between haploid nuclei. (F–H) Young spermatid nuclei presenting chromatin bridges (arrows). Some nuclei present very long chromatin threads (>50 µm; arrows in G). (I) A larger view of a testis with disorganized spermatid nuclei that harbor long chromatin threads. (J,K) Chromatin threads observed in spermatid nuclei that have incorporated protamines (arrows). Scale bars: 10 µm. (L) Quantification of abnormal spermatid nuclei in the indicated genotypes are represented as a scatter plot in which each point shows the number of nuclei with a chromatin thread counted in a single testis. Horizontal lines represent means. The loss of a single copy of K81 or hiphop in the Rescav background results in chromatin bridges at high frequency. Mann–Whitney test. n.s., non significant; ***P<0.001; ****P<0.0001. Note that abnormal spermatid nuclei in Rescav; K812/K812 testes were too numerous for reliable quantification (>300).