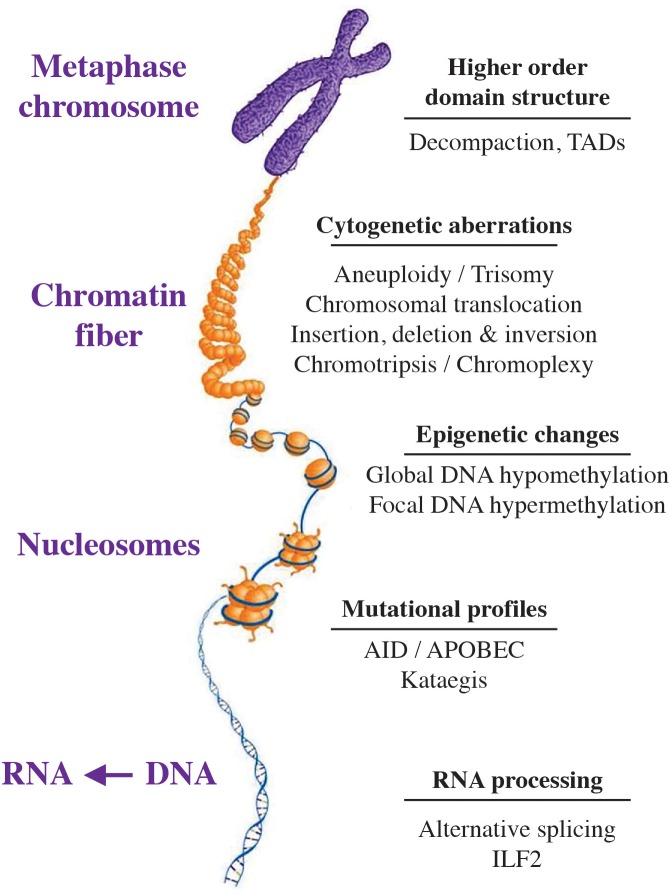
FIGURE 1.
Manifestation of genetic instability at all levels of the myeloma genome. The hierarchical organization of the genome at the chromosomal, chromatin fiber, nucleosomal and nucleotide level is indicated by a scheme that is labeled. Genomic changes commonly seen in myeloma are listed on the right. Recent findings indicate that myeloma exhibits substantial epigenetic change that relies on a small set of transcription factors, including members of the IRF (interferon regulatory factor), ETS (E26 transformation-specific), MEF2 (myocyte-specific enhancer factor 2), E-Box (enhancer box) and AP-1 (activator protein 1) families of proteins. Also included are E proteins, such as TCF3 (transcription factor 3) a.k.a. E2A (E2A immunoglobulin enhancer-binding factors E12/E47), TCF4 (transcription factor 4) a.k.a. ITF-2 (immunoglobulin transcription factor 2), and TCF12 (transcription factor 12) (Jin et al., 2018). Jin et al. (2018) also showed that de-compaction of heterochromatin is a defining feature of myeloma cells, which is in line with evidence that the myeloma genome undergoes genome-wide DNA hypo-methylation in the course of tumor progression (Agirre et al., 2015). AID, activation-induced cytosine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide; ILF2, interleukin enhancer binding factor 2; TADs, topologically associated domains.