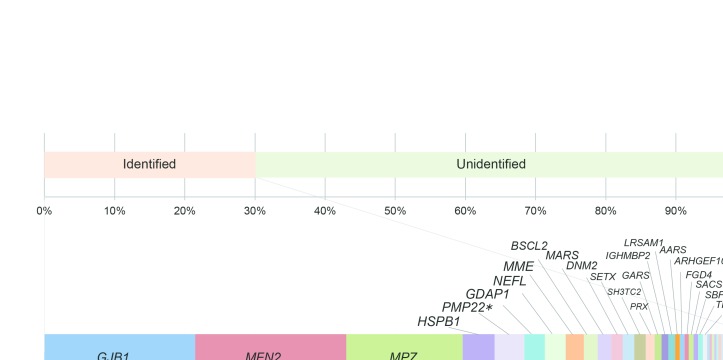
Figure 1.
Genetic spectrum of 301 cases with pathogenic or likely pathogenic variants. The following genes are indicated: GJB1 (21.9%), MFN2 (21.9%), MPZ (16.9%), HSPB1 (4.6%), PMP22 point mutation (4.3%), GDAP1 (3.0%), NEFL (3.0%), MME (2.7%), BSCL2 (2.0%), MARS (2.0%), DNM2 (1.7%), SETX (1.7%), SH3TC2 (1.7%), PRX (1.3%), GARS (1.0%), IGHMBP2 (1.0%), LRSAM1 (1.0%), AARS (0.7%), ARHGEF10 (0.7%), FGD4 (0.7%), SACS (0.7%), SBF2 (0.7%), TRPV4 (0.7%), TTR (0.7%), COA7 (0.3%), DCTN1 (0.3%), DHTKD1 (0.3%), EGR2 (0.3%), FBLN5 (0.3%), GALC (0.3%), GAN (0.3%), HARS (0.3%), HSPB3 (0.3%), HSPB8 (0.3%), INF2 (0.3%), KARS (0.3%), MTMR2 (0.3%), PRPS1 (0.3%), RAB7A (0.3%) and SOX10 (0.3%).