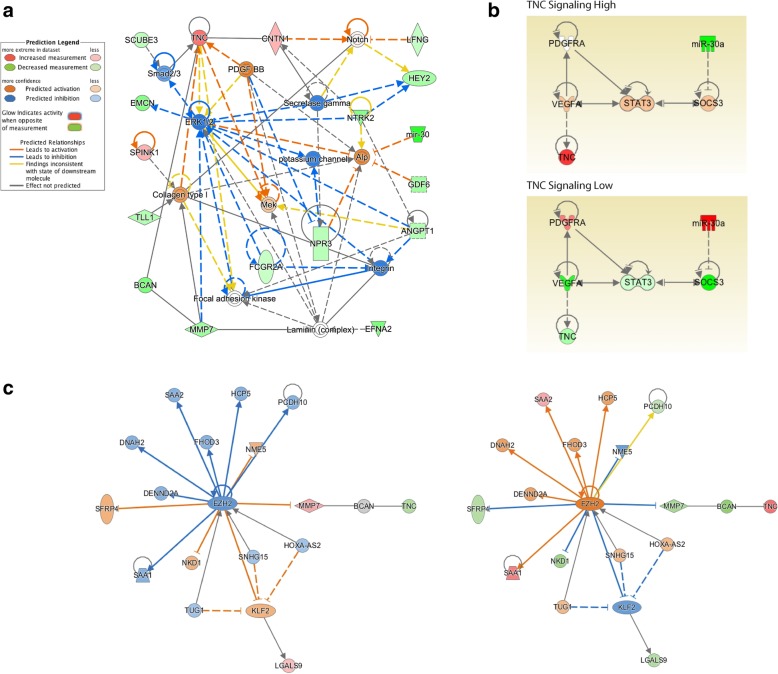
Fig. 6.
Functional pathways analysis reveals networks of molecular interaction and a potential mechanism of subgroup TNC overexpression in pediatric glioma
Pathways analysis was performed to evaluate the biological functions and interactions of differentially expressed genes between TNC cDNA and shRNA cell lines (Log2Fold Change > 2 or < − 2, adjusted p-value < 0.05). a Organismal Development and Small Molecule Biochemistry is indicated as the top network of molecular interaction (22 focus molecules, score 45), with TNC over expression, miRNA30a downregulation, PDGF and NOTCH signaling activation, and interaction with extracellular matrix proteins. These results suggest that the observed biological effects of elevated TNC expression in vitro may be due to these pathway activations. b VEGFRA and STAT3 activation, potentially affected by miRNA30 expression level, are identified as potential upstream regulators of TNC overexpression in a subgroup of DIPGs lacking PDGFRA amplification or mutation. c EZH2 is implicated as a top activated upstream regulator in TNC cDNA (left) relative to TNC shRNA (right) cell lines (activation z-score 1.639, p-value of overlap 0.004), with a direct downstream effect on TNC expression