Figure 1.
Quantitative Temporal Analysis of VACV Infection
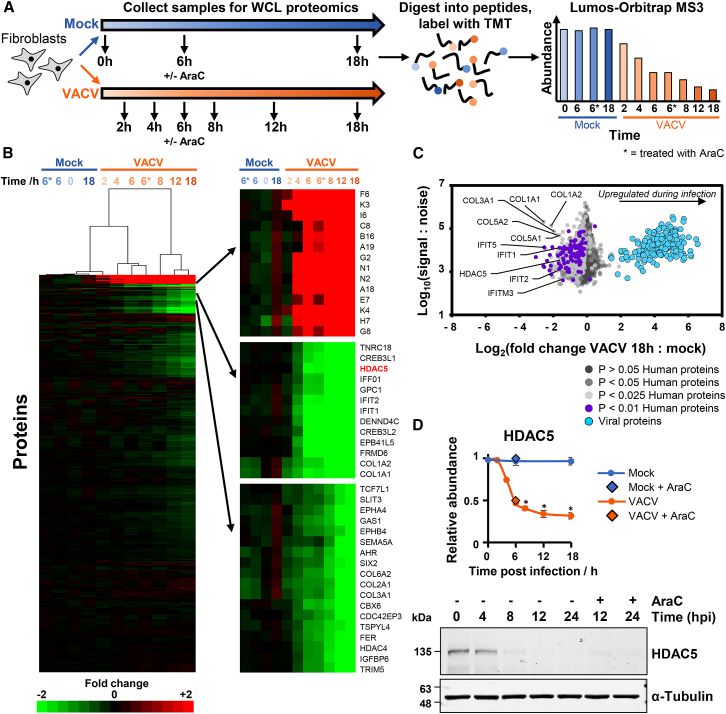
(A) Schematic of experimental workflow for each of three biological replicates. Cells were infected at MOI of 5 or mock infected (Figure S1A). Additionally, one mock and one infected sample were treated for 6 h with the viral DNA replication inhibitor cytosine arabinoside (AraC).
(B) Hierarchical cluster analysis of all proteins quantified. An enlargement of three subclusters is shown (right panel), including multiple proteins that were substantially up- or downregulated.
(C) Scatterplot of all proteins quantified at 18 h of infection. For all analyses in this manuscript, a mean fold change at each time point was calculated by averaging fold changes from each of the biological replicates in which the protein was quantified. For the purposes of comparison, the 18-h mock sample from each replicate was used, because the 0-, 6-, and 18-h mock samples behaved extremely similarly (Figure S1E). To perform a comprehensive analysis, “sensitive” criteria were employed, examining proteins down- or upregulated >2-fold on average across all replicates in which the protein was quantified (Table S2A). Sensitive criteria were used in each analysis apart from where indicated. Data from “stringent” criteria that examined only proteins quantified in all three replicates with an average fold change > 2 and p < 0.05 are shown in Table S2C. For proteins quantified in all three replicates, a Benjamini-Hochberg-corrected two-tailed t test was used to estimate p values. Only proteins quantified in all three replicates are shown in this scatterplot.
(D) Example of a previously unreported target of VACV infection. Data are represented as mean ± SEM; ∗p < 0.05 (see STAR Methods). Immunoblot of HFFF-TERTs infected with VACV (MOI = 5) confirmed rapid HDAC5 downregulation.