Fig. 6.
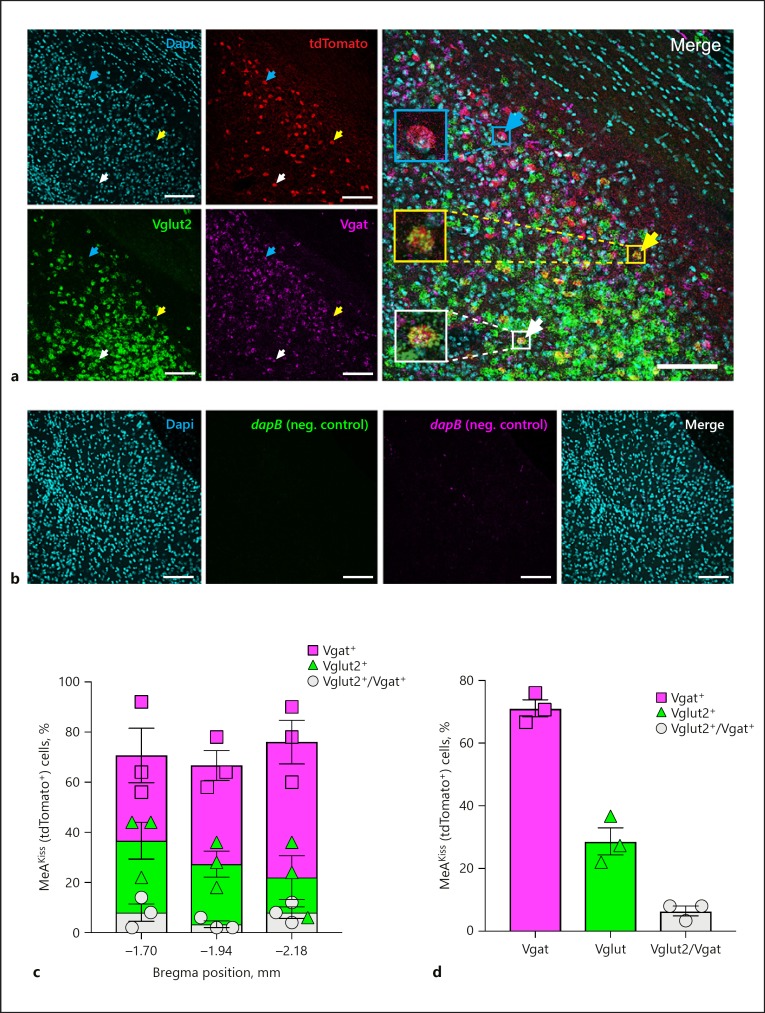
Amygdala Kiss1 neurons express mRNAs encoding Vgat and Vglut2 in male mice. a A representative confocal micrograph (16 µm thickness) displays Dapi nuclear stain (cyan), tdTomato (red), Vglut2 (green), and Vgat (magenta) expression in the MeA in Kiss1CreEGFP/wt/Rosa26lox-stop-lox-tdTomato/wt male mice. A cyan arrow marks a tdTomato+ cell that expresses Vgat mRNA, a yellow arrow marks a tdTomato+ cell that expresses Vglut2 mRNA, and a white arrow marks a tdTomato+ cell that expresses both. Scale bars, 100 µm. b Representative confocal micrographs (16 µm thickness) that display negligible RNAscope in situ signals with RNAscope 3-plex negative control probes in the MeA in Kiss1CreEGFP/wt/Rosa26lox-stop-lox-tdTomato/wt male mice. Dapi nuclear stain (cyan); RNAscope 3-plex negative control probe (dapB; a bacterial gene) developed in the FITC channel (green); RNAscope 3-plex negative control probe (dapB; a bacterial gene) developed in the Cy5 channel (magenta), and Merge of all three channels. Scale bars, 100 µm. c Quantification of labelled MeAKiss neurons expressing Vgat (squares), Vglut2 (triangles), or both (circles) as a percentage of tdTomato+ cells at three Bregma positions (n = 150 cells monitored in each of 3 animals). d The overall percentage of labelled MeAKiss neurons expressing Vgat (squares), Vglut2 (triangles), or both (circles) (n = 3). Data are represented as mean ± SEM with the value from each individual mouse indicated.