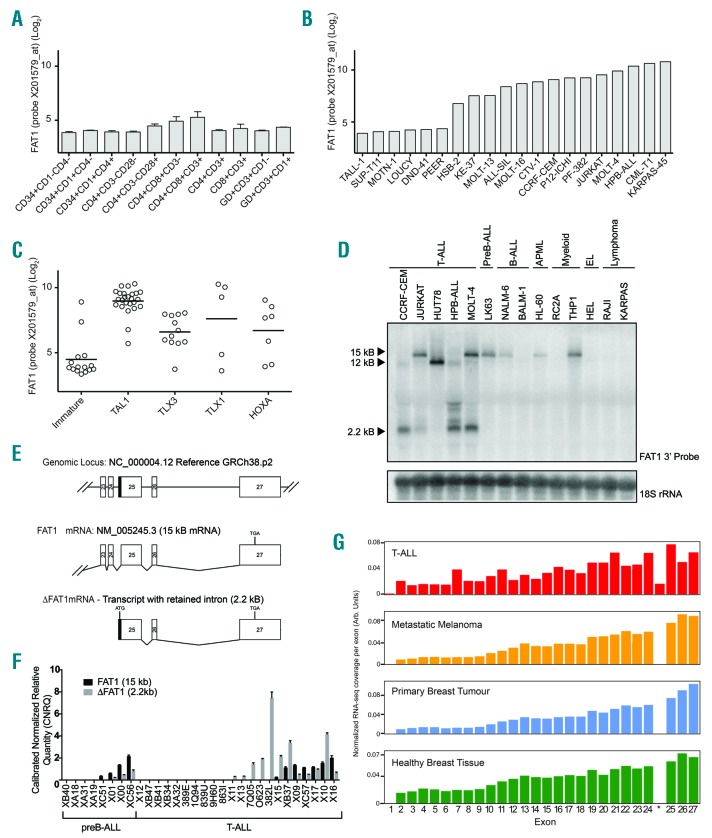
Figure 1.
Truncated FAT1 transcript with a retained intron is uniquely expressed in T-ALL. (A) FAT1 is expressed at low levels across a diverse range of normal T-cell subsets. (B) In contrast 14/20 T-ALL cell lines expressed high levels of FAT1 transcript. (C) In clinical T-ALL cases, FAT1 is expressed predominantly within TAL1-positive cases and is negatively correlated with immature cases of T-ALL. (D) Northern blot analysis of T-ALL cell lines identified multiple 5’ truncated FAT1 transcripts present with the majority expressing a band migrating at 2.2 kb. (E) Schematic representation of the 3’ exon structure of the FAT1 gene locus and the 5’ RACE result showing that the 2.2 kb truncated transcript contains a retained intronic sequence directly upstream of exon 25. (F) Real-time polymerase chain reaction analysis of B- and T-ALL patient-derived xenograft samples shows that full-length FAT1 is expressed in both TALL and B-ALL cases but the ΔFAT1 transcript is present at high levels only in T-ALL cases. (G) Analysis of the TCGA and TARGET databases revealed that the retained intronic sequence denoted by * is unique to T-ALL cases when compared to melanoma and breast cancers that also have high levels of FAT1 expression.