Figure 7.
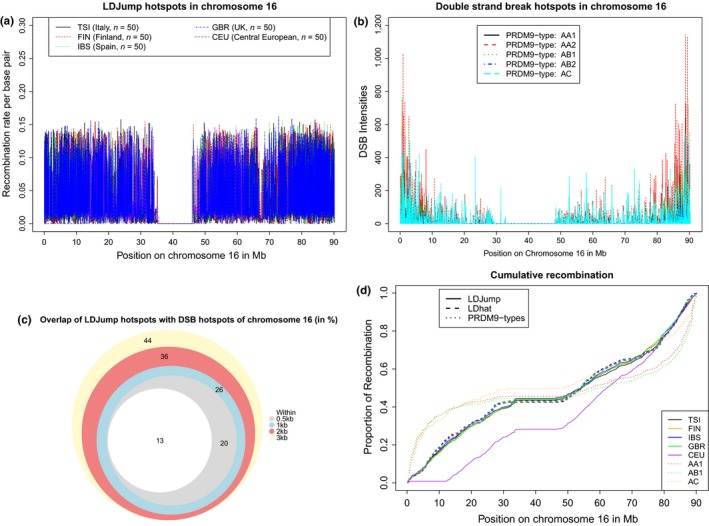
(a) Estimated recombination map for chromosome 16 of four European populations with 50 randomly sampled sequences of the 1000 Genomes Project using LDJump under the demography model and segment lengths of 1 kb. The results of the Italian sample are plotted in black, the Finnish sample in dashed red, the Spanish population in dotted green and the British one in dash‐dotted blue. (b) Double‐strand break maps taken from Pratto et al. (2014) of chromosome 16 for five individuals with different PRDM9‐types. Here, the different colours and line types represent different individuals (AA1, solid‐black; AA2, dashed‐red; AB1, dotted‐green; AB2, dash‐dotted‐blue; AC, long‐dashed‐cyan). (c) Overlap between detected DSB hotspots and the hotspots identified by LDJump. With LDJump, we define hotspots as regions with more than five times the estimated background rate. The DSB hotspots were taken from Pratto et al. (2014). We looked at overlaps between LDJump and the DSB hotspots that occurred with at least one European population (white areas). To assess the level of accuracy, we added segments of length 0.5 (grey), 1 (cyan), 2 (red), and 3 (yellow) kb left and right to the DSB region boundaries. The comparison is performed for all PRDM9‐types considered in Pratto et al. (2014). The total number of DSB hotspots of all PRDM9‐types is 2889 of which 866 were not within 3 kb of a LDJump hotspot. (d) Accumulation of recombination within chromosome 16 with increasing sequence for each method and population estimated with LDJump (solid lines), LDhat maps from the 1000G (dashed lines), DSB intensities for three individuals (dotted lines) (Pratto et al., 2014). The colour coding remains the same for the five European populations. The shift in cumulative recombination of the CEU compared to the other populations results from a lack of information up to position 10 MB [Colour figure can be viewed at wileyonlinelibrary.com]
