Figure 1.
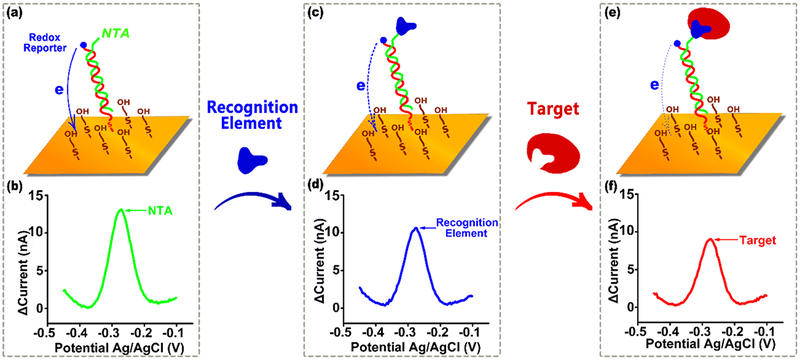
E-DNA scaffold sensors are composed of a redox-reporter-modified, double stranded nucleic acid “scaffold” attached via a flexible linker to self-assembled monolayer deposited on a gold electrode. Their distal end is modified with a redox-reporter (here methylene blue) and a target-binding recognition element. Here we have attached these recognition elements using a nitrilotriacetic acid (NTA) on the scaffold that, in the presence of copper, binds to a His-tag on the recognition element. (a) In the absence of a recognition element the reporter readily approaches the electrode, (b) producing a large redox current. (c) Upon addition of a recognition element (during sensor fabrication) this current is (d) reduced, presumably due to the steric bulk of the recognition element. (e) The binding of a macromolecular target, such as an antibody, (f) reduces this current still further (Figure S1), producing an easily measurable signal indicative of the presence of the target. Here we explore how large a recognition element can be employed before the reduction in current it causes is so great that target binding no longer produces a sufficient additional signal change to support target detection.