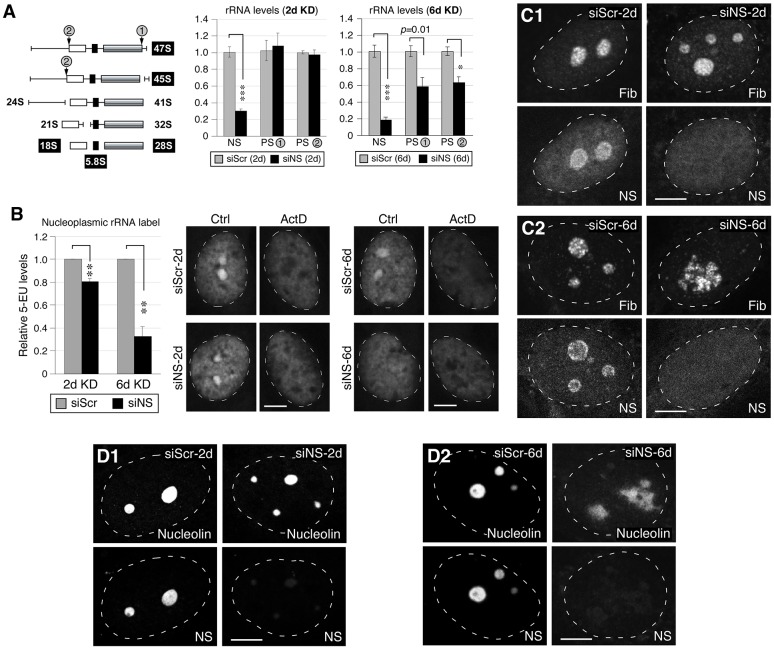
Fig. 3.
Knockdown of nucleostemin has minimal effects on rRNA synthesis or nucleolar integrity at the 48-hour time-point. (A) Left, diagram showing the pre-rRNA-processing step and the primer positions flanking the processing sites 1 and 2 (PS-1, PS-2). White, black and grey boxes represent 18S, 5.8S and 28S rRNAs, respectively. The PS-1 and PS-2 primer recognition sites are indicated by gray circles. Middle and left panels, qRT-PCR assays of nucleostemin (NS), PS-1 and PS-2 levels in MDA-MB-231 cells treated with siScr or siNS for 2 days (2d) or 6 days (6d). The expression levels of target transcripts were quantified by normalizing to the expression of HMG-14. The expression levels of each transcript in the corresponding siScr samples were arbitrarily set as 1. (B) A modified 5-EU click-chemistry assay was designed to measure the amount of newly synthesized mature rRNA present in the nucleoplasm. The differential 5-EU signals [control (Ctrl) minus actinomycin (ActD)-treated samples] in the nucleoplasm were quantified in MDA-MB-231 cells treated with siScr or siNS for 2 days or 6 days. Data show the mean±s.e.m.; *P<0.01, **P<0.001, ***P<0.0001. (C) Confocal immunofluorescence of the fibrillarin (Fib)-labeled dense fibrillar component and nucleostemin-labeled structure in siRNA-treated cells at the 2-day and 6-day time-points. (D) Confocal immunofluorescence of nucleolin- and nucleostemin-labeled nucleolar structures in siRNA-treated cells at the 2-day and 6-day time-points. White dashed lines show nucleus outlines. Scale bars: 10 µm.