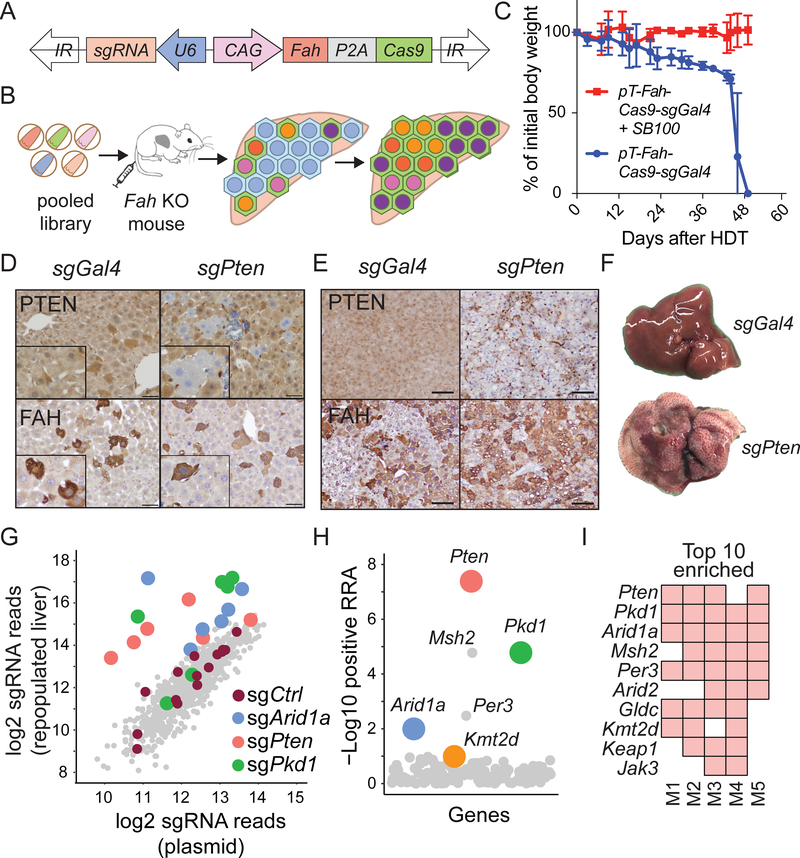
Figure 5. In-vivo CRISPR screening identified genes that increase clonal expansion.
A. Sleeping Beauty transposon used for stable hepatocyte expression of FAH, Cas9, and sgRNA. The plasmid was injected intravenously.
B. Schema of in vivo loss-of-function screen to identify genes that regulate liver regeneration using the Fah KO hereditary tyrosinemia model.
C. Body weights of Fah KO mice undergoing liver repopulation after plasmids were delivered +/− SB100 transposase plasmid by hydrodynamic transfection (HDT). NTBC was withdrawn immediately after HDT. Data are represented as mean ± standard deviation; n = 3 mice per group.
D. FAH IHC staining shows FAH+, PTEN negative hepatocytes one week after HDT of plasmids carrying Fah, Cas9, and a Pten sgRNA (scale bar = 50 μm).
E. Representative IHC staining one month after transposon HDT (scale bar = 100μm).
F. The appearance of livers 1 month after transposon HDT.
G. Scatterplot showing average enrichment of individual sgRNAs after liver repopulation from 5 independent replicates. The sgRNA count was defined as the number of sequencing reads that perfectly match the sgRNA target sequence (see Supplemental Table 5).
H. Identification of candidate genes using positive robust rank aggregation (RRA) score as assessed by MAGeCK. The RRA score reflected whether or not the distribution of sgRNAs targeting a gene were significantly skewed within a ranked list of all sgRNAs. Assuming that if a gene had no biological effect, then sgRNAs targeting this gene should be uniformly distributed (Li et al., 2014).
I. Top 10 gene candidates in the repopulated liver based on RRA scores. The screen was performed in 5 independent mouse replicates. A red square means that the gene was found within the top 10 enriched genes in that screened mouse.