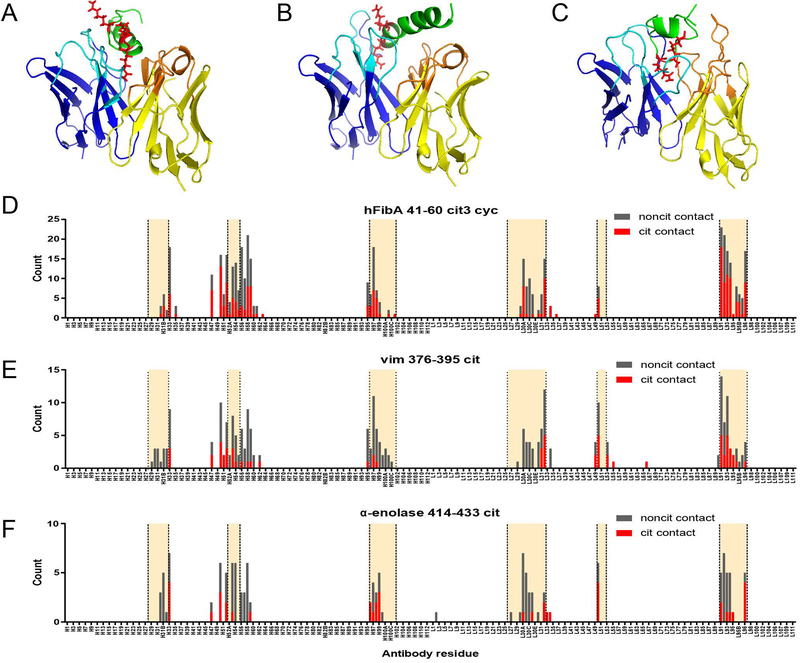
Figure 5.
Predicted contact residues of ACPA clonal family antibodies based on molecular modeling. Molecular models of antibodies from clonal families 1, 2, and 3 from Fig. 4 were producing using the Rosetta software package. Three peptides, hFibA 41–60 cit3 cyc (A, D), vim 376–395 cit (B, E), and α-enolase cit2 (C, F), were modelled using PEP-FOLD 2.0 and converted to citrullinated forms using PyTM. Antibody-antigen docking was performed using Rosetta (see Methods). Three representative molecular models of patient-derived mAbs docked with citrullinated peptides are presented: rMab 80 docked with hFibA 41–60 cit3 cyc peptide (A), rMab 78 with vim 376–395 cit peptide (B), and rMab 66 with α-enolase 414–433 cit2 peptide (C). The predicted contact residues for each docked model were counted and the cumulative results of all antibody-antigen combinations is presented (D-F). Antibody sequences are presented using the Chothia (post-1997) numbering scheme with the heavy and light chains concatenated along the X-axis. CDR regions are highlighted between the vertical dotted lines.