Extended Data Figure 5. |. Efficacy of genetic and pharmacological interventions.
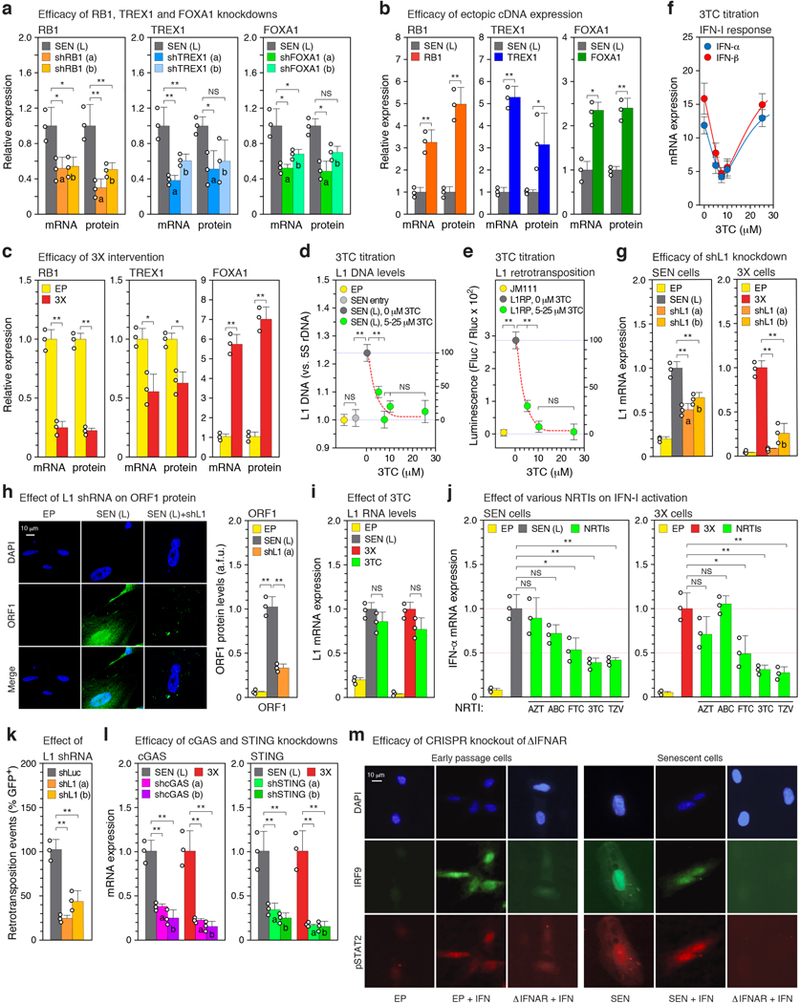
a, Knockdowns with two distinct shRNAs (a, b) or b, ectopic cDNA expression were performed in senescent cells as described in Fig. 2d, e, g (also see Methods). The effectiveness of these manipulations on their targets was assessed by RT-qPCR and immunoblotting. For gel source data see Supplementary Fig. 1. c, RB1, TREX1 and FOXA1 mRNA and protein expression after the triple (3X) intervention (Fig. 2f). d, The effect of 3TC treatment on the relative abundance of L1HS sequences in senescent cells was determined by multiplex TaqMan qPCR on total DNA (primer set 6, Supplementary Table 1). SEN entry, 0 weeks in senescence (Fig. 1a; point A in Extended Data Fig. 1a). 3TC was administered continuously from SEN entry until harvest 16 weeks later. e, The dual luciferase L1 reporter system52 was used to determine the effect of 3TC dosing on retrotransposition. L1 reporters were introduced into early passage cells using lentivirus vectors (Methods) and cells were treated with 3TC for 4 days prior to harvest and assay. JM111, a defective reporter carrying mutations in ORF1 (absence of 3TC); L1RP, a retrotransposition competent reporter. f, The effect of 3TC dosing on the IFN-I response. The experiment above (d) was processed by RT-qPCR to determine the expression of IFN-α and IFN-β1. g, Knockdowns of L1 were performed with two distinct shRNAs (a, b) in senescent cells (as in Fig. 2d, e, g) or 3X cells (as in Fig. 2g). The effectiveness on L1 expression was assessed by RT-qPCR using poly(A)-purified RNA and primers F. h, Cells in the experiment in (g) were examined for levels of ORF1 protein by immunofluorescence (IF). Image analysis was performed with CellProfiler software (Methods). >200 cells were examined for each condition (a.f.u., arbitrary fluorescence units). i, The L1 shRNA treatment in the experiment in (g) was substituted with 3TC treatment (10 μM) for the same period of time. j, Five different NRTIs (or combinations) were tested for effects on the IFN-I response. AZT (Zidovudine, 15 μM), ABC (Abacavir, 15 μM), FTC (Emtricitabine, 10 μM), 3TC, (Lamivudine, 10 μM), TZV (Trizivir, a combination of 15 μM AZT, 15 μM ABC and 7.5 μM 3TC). Cells were treated for 4 weeks between 12 and 16 weeks in senescence (Fig. 1a; points D and E in Extended Data Fig. 1a). 3X cells (Fig. 2f) were treated with 3TC for 48 hours after the completion of the last drug selection. Interferon α expression was determined by RT-qPCR. k, A native L1 reporter (pLD143)53 was co-transfected with shRNA plasmid vectors into HeLa cells (Methods). Retrotransposition was scored as GFP-positive cells, and shL1 knockdowns were normalized to a shLuc negative control. The absolute average retrotransposition frequency (percentage of GFP-positive cells) was 4.1, which matches the published values for the reporter used (pLD143)53. l, Knockdowns of cGAS and STING were performed in senescent or 3X cells as with the other shRNAs (Fig. 2d, e, g and a, g above). m, Downregulation of interferon signaling after CRISPR-mediated inactivation of IFNAR1 and IFNAR2 genes was verified by the absence of IRF9 nuclear translocation and STAT2 phosphorylation in response to interferon stimulation. Cells were infected with lentivirus vectors expressing Cas9 and gRNAs to both IFNAR1 and IFNAR2 (ΔIFNAR, Methods). After the infection cells were re-seeded on coverslips, treated with interferon for 2 hours, and examined by IF microscopy. The experiment was repeated 3 times with similar results. (a-i, l) n = 3 independent experiments. (k) n = 3 independent biological samples, repeated in 2 independent experiments. (a-l) Data are mean ±s.d. *P ≤ 0.05, **P ≤ 0.01, unpaired two-sided t-tests. Exact P values can be found in the accompanying Source Data.
