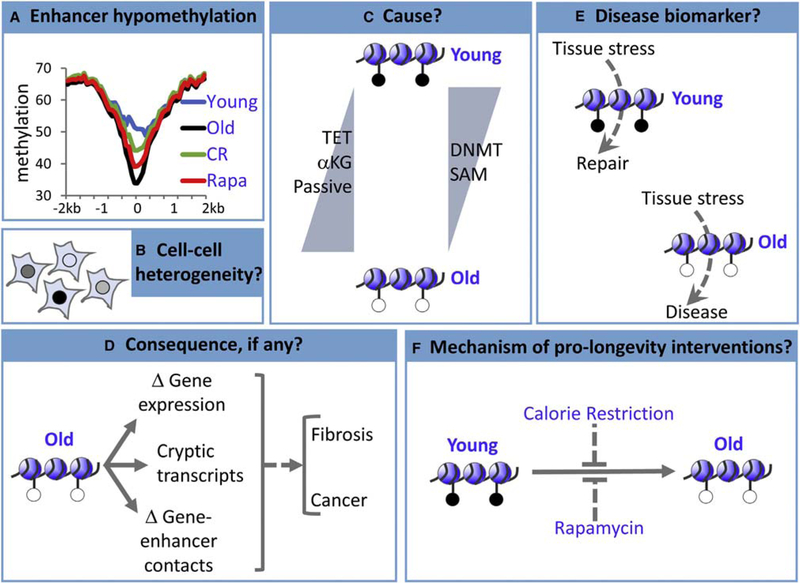
Figure 5. Some Outstanding Questions.
(A) In the mouse liver, hypomethylation of enhancers is linked to chronological age and contributes to the clock, but the rate of hypomethylation is decreased by interventions thought to slow biological aging (calorie restriction (CR), rapamycin (rapa), and Ames dwarfism [not shown]), suggesting that enhancers are a hybrid chronological and biological clock (but potentially separable) (Cole et al„ 2017; Wang et al., 2017).
(B-F) Many questions remain, although the answers to these questions likely differ for different clock CpGs (e.g., at enhancers versus promoters).
(B) To date, methylation clocks have been generated from data obtained from populations of cells, is there cell-to-cell variation in ticking of the clock?
(C) Does enhancer hypomethylation result from downregulation of DNMTs or their methyl donor substrate (SAM), increased TET demethylase activity, increased TET cofactor α-KG, increased passive demethylation, or another mechanism?
(D) At enhancers, does hypomethylation affect gene expression, expression of cryptic transcripts that are normally silenced by DNA methylation, enhancer-gene interactions, or all or none of these?
(E) Can a biological clock, probably tissue-specific, predict disease with sufficient sensitivity and specificity to be clinically useful?
(F) How do pro-longevity interventions slow ticking of the clock, and does this contribute to their pro-longevity and/or pro-health aging benefits?