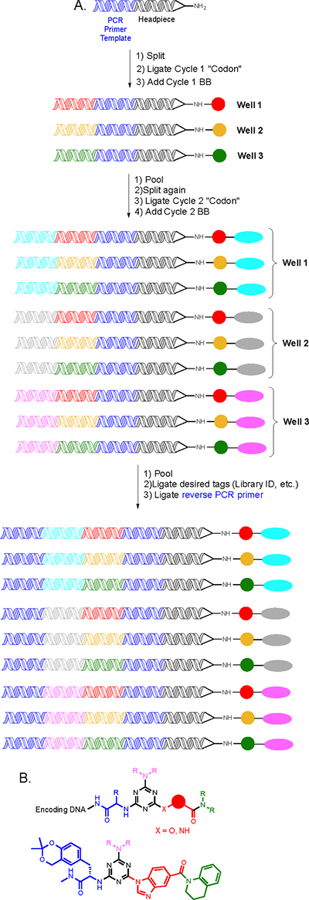
Figure 1. Split and pool synthesis of a DNA-encoded library.
A. Schematic depiction of a two cycle DEL using three building blocks at each position to create nine unique species. In general, the size of libraries created by split and pool chemistry will be xy, where x = the number of unique building blocks used at that position and y = the number of cycles used to create the library. B. The general structure of an 800 million compound library containing a central triazine core. The structure of a screening hit against p38 MAP kinase (the identity of the unit shown in pink was not relevant to binding).