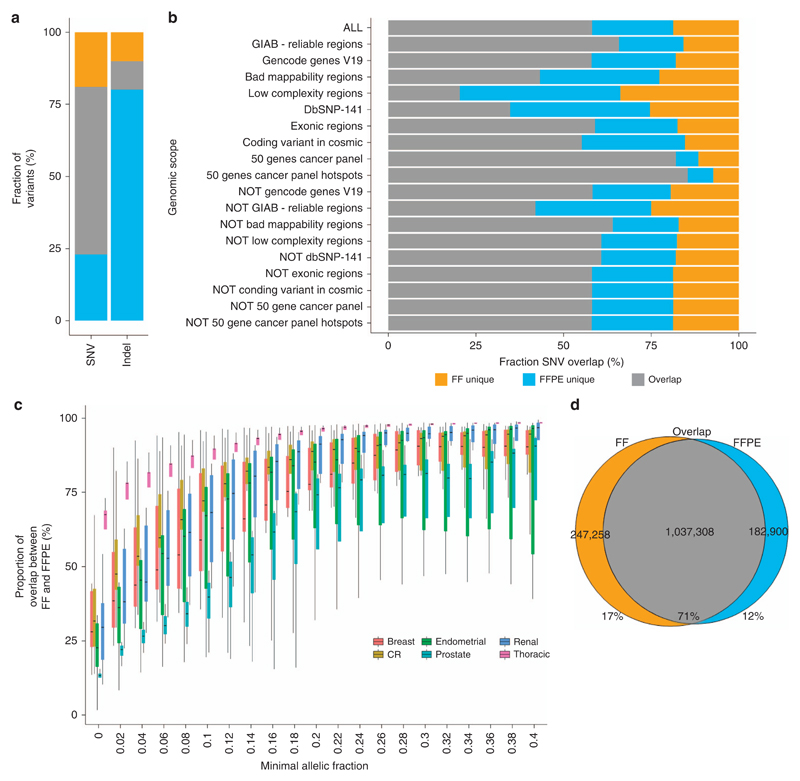
Figure 2. Somatic SNVs and small insertions and deletions detection and their respective overlap between FF and FFPE samples.
(a) Agreement between FF and FFPE samples of somatic SNVs and indels represented in percent. The three sections represent the FF-unique, FFPE-unique, and FF–FFPE overlap variants. (b) Overlap between FF and FFPE samples of somatic SNVs in different scopes of the genome showing the fraction of variant unique in FF, unique in FFPE, and in both FF and FFPE (the different regions are detailed in Supplementary Table S8). (c) Evolution of the proportion of SNVs in common in FF and FFPE samples when variants with low allelic fractions are filtered from the data set for each tissue type: renal (N = 14), CR (N = 12), prostate (N = 4), breast (N = 10), endometrial (N = 7), and thoracic (N = 5). (d) Agreement in SNVs between FF and FFPE samples. The following variants were considered for this plot: variants in reliable regions (Genome in a Bottle regions) where the depth needed to be at least 70 × at the position of the variant for both FF and FFPE samples and the allelic fraction needed to be at least 0.067 in one of the two samples (see Supplementary Figures S12 and S19 for details). CR, colorectal; FF, fresh-frozen sample; FFPE, formalin-fixed, paraffin-embedded sample; GIAB, Genome in a Bottle; SNV, single-nucleotide variant.