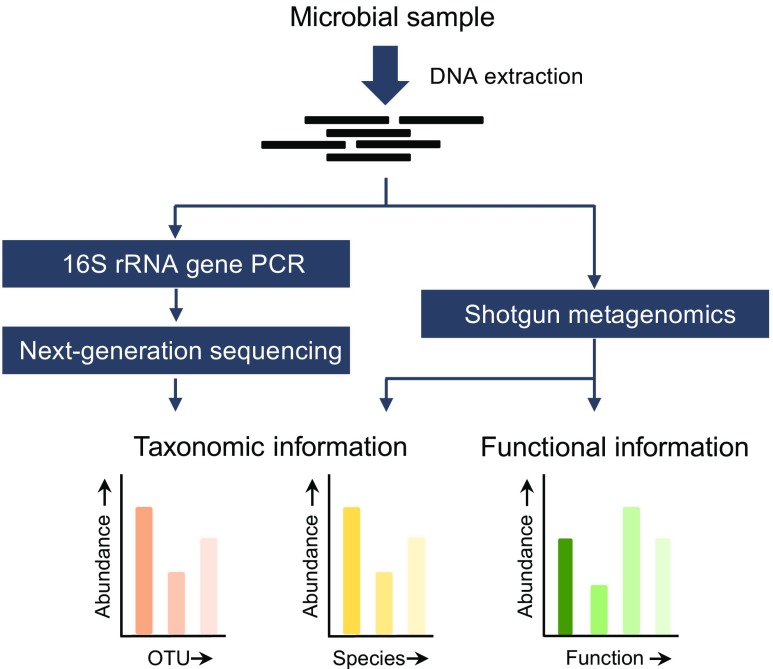
Fig. 1.
General overview of 16S rRNA gene NGS and shotgun metagenomics methods. Both methods start with the extraction of nucleic acids from a microbial sample. Next, the extracted DNA is either subjected to 16S rRNA gene PCR amplification (16S rRNA gene NGS) or sheared into small DNA fragments (shotgun metagenomics). The resultant 16S rRNA gene amplicons, or sheared DNA fragments, are sequenced using NGS techniques. Finally, all sequence data are processed using an extensive array of bioinformatics algorithms that allows the researcher to explore the taxonomic composition and/or the functional capacity of the sample tested. OTU operational taxonomic units—a group of very similar sequences.