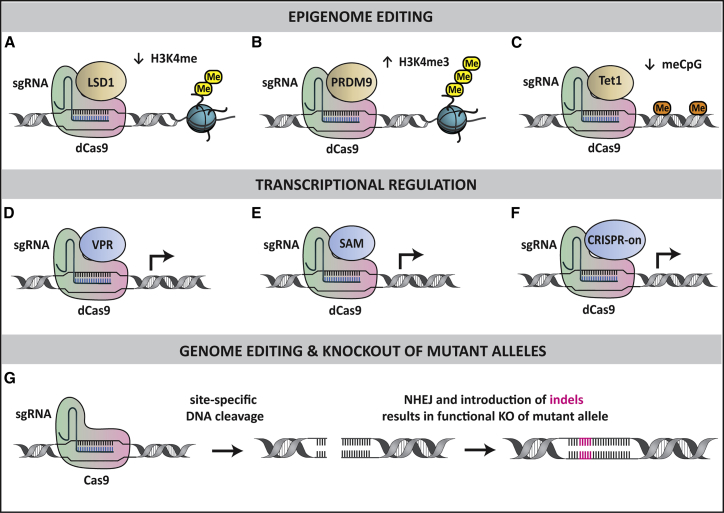
Figure 4.
Epigenome Editing, Transcriptional Regulation, and Genome Editing with CRISPR-Cas9
Epigenome editing. dCas9-sgRNA complexes can be fused to epigenetic enzymes, such as (A) lysine-specific histone demethylase 1 (LSD1), (B) histone-lysine N-methyltransferase PRDM9, and (C) methylcytosine dioxygenase Tet1, for targeted epigenome editing at specific DNA loci. Transcriptional regulation. dCas9-sgRNA complexes can also be fused to engineered transcriptional regulatory complexes, such as (D) VP64-p65-Rta (VPR), (E) Synergistic Activation Mediator (SAM), and (F) CRISPR-on, to activate gene expression at specific loci. Genome editing and knockout of mutant alleles. (G) Cas9-sgRNA complexes generate a double-stranded break at the targeted allele. In mitotically quiescent cells, such as SCs of the mature mammalian cochlea, breaks can be repaired by the non-homologous end joining (NHEJ) process that leaves small insertions and deletions (indels) at the cut site. This produces frameshift and nonsense mutations, resulting in functional knockout (KO) of the mutant allele.