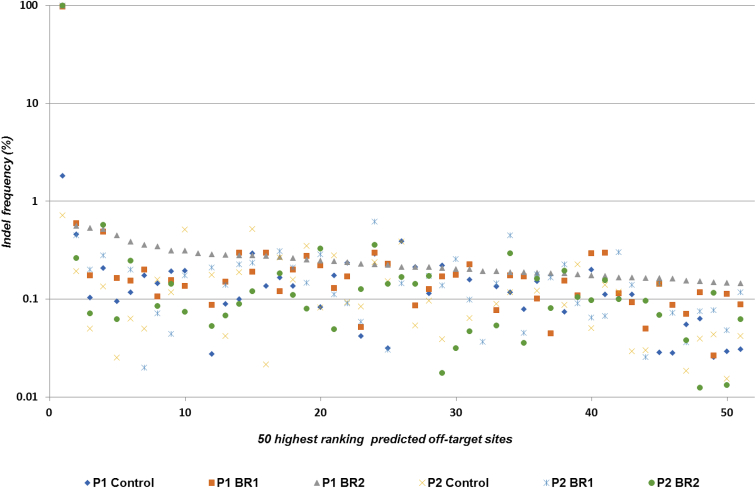
Figure 3.
NGS Assessment for the Presence of Indels in Potential Off-Target Sites
PCR amplicons for 244 predicted off-target sites sequenced with at least 1,000× depth, as well as the on-target region, were analyzed (x axis, putative off-target sites arranged according to the frequency of sequence variation). The 50 highest ranking sites are represented. The first point corresponds to the on-target site (y axis, percentage of reads containing sequence variations within a 6-bp window centered on the target site for each sgRNA).