Abstract
Background and Aims
Non-dividing hepatocytes in end-stage liver disease indicates permanent growth arrest similar to senescence. Identifying senescence in vivo is often challenging and mechanisms inhibiting senescence are poorly understood. In lower organisms mitochondrial unfolded protein response (UPRMT) helps in increasing longevity; however, its role in senescence and liver disease is poorly understood. Aim of this study was to identify hepatocyte senescence and the role of UPRMT in cryptogenic cirrhosis.
Methods
Doxorubicin was used to induce senescence in non-neoplastic hepatocytes (PH5CH8) and hepatoma cells (HepG2 and Huh7). Senescence-associated markers and unfolded protein response was evaluated by fluorescence microscopy, immunoblotting and gene expression. Explants/biopsies from normal, fibrosis, compensated and decompensated cirrhosis without any known etiology were examined for presence of senescence and UPRMT by immunohistochemistry and gene expression.
Results
Accumulation of senescent hepatocytes in cryptogenic cirrhosis was associated with reduced proliferation, increased expression of γH2AX and p21, together with loss of LaminB1. Dysfunctional mitochondria and compromised UPRMT were key features of senescent hepatocytes both in vitro and also in decompensated cirrhosis. Intriguingly, compensated cirrhotic liver mounted strong UPRMT, with high levels of mitochondrial protease, CLPP. Overexpression of CLPP inhibited senescence in vitro, by reducing mitochondrial ROS and altering oxygen consumption.
Conclusions
Our results implicate a role of hepatocyte senescence in cryptogenic cirrhosis together with a crucial role of UPRMT in preventing hepatocyte senescence. A compromised UPRMT may shift the fate of cirrhotic liver toward decompensation by exaggerating hepatocyte senescence. Restoring CLPP levels at least in cell culture appears as a promising strategy in mitohormesis, thereby, preventing senescence and possibly improving hepatocyte function.
Keywords: Cryptogenic Liver Cirrhosis, Mitochondrial Respiration, Mitochondrial Unfolded Protein Response, Oxidative Stress
Abbreviations used in this paper: cDNA, complementary DNA; CLPP, Caseinolytic mitochondrial matrix peptidase proteolytic subunit; Dox, doxorubicin; GFP, green fluorescent protein; IHC, immunohistochemistry; mtDNA, mitochondrial DNA; nDNA, nuclear DNA; OCR, oxygen consumption rate; qPCR, quantitative polymerase chain reaction; SA-β-gal, senescence-associated β-galactosidase; TMRE, tetramethylrhodamine ethyl ester; UPRMT, mitochondrial unfolded protein response
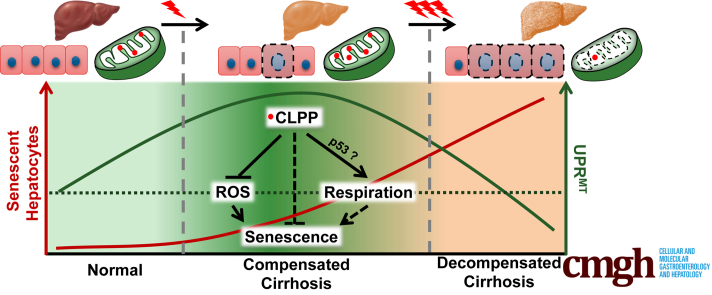
Graphical abstract
Summary.
Compensated cirrhosis exhibited a marked increase in mitochondrial unfolded protein response with the high levels of CLPP. However, decompensated cirrhosis showed reduced UPRMT with a significant increase in number of senescent hepatocytes. Overexpression of CLPP in an in vitro model of premature senescence could significantly reduce senescence-associated phenotype by inhibiting mitochondrial ROS and altering mitochondrial respiration.
Liver injury usually leads to high turnover of cells which includes both cellular death and regeneration. During chronic liver injury, impairment in regenerative capacity results in functional insufficiency, often culminating in cirrhosis, which is also clinically regarded as end-stage liver disease. Two distinct subclinical stages have been proposed for cirrhosis which includes (1) an early compensated phase with or without varices and (2) a late decompensated phase with life threatening complications such as variceal hemorrhage, ascites, upper gastrointestinal bleed, or hepatic encephalopathy.1 The evolution of cirrhosis involves a pathological transition from highly regenerating nodules in compensated state to explicatively exhausted hepatocytes during decompensated state, finally culminating in parenchymal extinction.1 There are a few questions still unanswered regarding the cirrhotic evolution: what tips the balance in favor of a decompensated state and what mechanisms rescues hepatocytes from growth inhibition? In chronic liver disease, hepatocyte damage leading to cell death has been extensively studied, while the process of cellular senescence that prevents hepatocyte proliferation is relatively less explored. A senescent hepatocyte is permanently growth arrested, which inhibits liver regeneration and contributes to disease progression. Evidence now strongly points toward existence of senescent hepatocytes in various liver diseases related to alcohol, hepatitis C virus, and fatty liver.2, 3, 4 Mechanisms involved in hepatocyte senescence are poorly understood. Some studies have highlighted the role of cell cycle inhibitors like p21, transforming growth factor beta and telomere shortening in hepatocyte growth arrest and senescence.2, 5, 6, 7 Cellular senescence due to telomere attrition is usually a slow process; however, oxidative damage usually results in accelerated aging and often referred as stress-induced premature senescence. Oxidative damage is often a hallmark feature in cirrhotic liver,8, 9 which in turn implicates its role in genesis of senescent hepatocytes.
Mitochondria are the main sources of reactive oxygen species (ROS) and considered a main cause of aging.10 Hepatocytes are rich in mitochondria and altered mitochondrial functions have been documented in a variety of chronic liver diseases.11 Maintaining the mitochondrial function and removal of defective mitochondria are therefore important in preventing senescent changes and in maintenance of organ homeostasis. Dysfunctional mitochondria are removed by the process of mitophagy, which is often defective in senescence.12 Defective mitophagy has been well studied in the context of liver disease.13, 14 Recent studies have pointed to the existence of yet another cellular mechanism, named mitochondrial unfolded protein response (UPRMT) involving mitochondrial-to-nuclear communication or retrograde response for recovery of dysfunctional mitochondria.15 This protein quality control mechanism is well studied in model organisms, such as yeast, Caenorhabditis elegans, Drosophila melanogaster, etc. Work in C. elegans has revealed a link between UPRMT and enhanced longevity.16 This in turn implicates a role of UPRMT during aging including senescence. However, the role of UPRMT in the context of mammalian senescence is not well studied. As senescence is a stress response, it is essential to evaluate the role of UPRMT in this process. Senescent cells often accumulate in disease conditions, such as cirrhosis; there are hardly any data available on relevance of UPRMT in end-stage liver disease. Recently, 2 papers have highlighted contradictory roles of UPRMT in the liver. Gariani et al17 reported that nicotinamide adenine dinucleotide replenishment promoted UPRMT to prevent fatty liver. On the other hand, deletion of mitochondrial protease, CLPP, a key player of UPRMT, protected mice from development of fatty liver when fed on high-fat diet.18, 19
Identifying senescence in clinical specimens is often challenging and mechanisms involved in hepatocyte senescence are poorly understood. Further, strategies averting hepatocyte growth inhibition due to senescence appears crucial in preventing liver disease. As mitochondrial dysfunctions accompany liver disease, we hypothesized that alterations in mitochondrial stress response pathway (ie, UPRMT) may accompany senescent-associated changes during progression of liver disease and key players of UPRMT can ameliorate hepatocyte senescence. The aim of the present study was to identify senescence-associated markers together with alterations in UPRMT pathway using, first, an in vitro model of doxorubicin (Dox)-induced hepatocyte senescence and, second, during progression of end-stage liver disease in cryptogenic liver disease. There is hardly any information available on the molecular events associated with development of cryptogenic liver disease. Also, other forms of basic insults, such as alcohol, viruses, or fatty liver disease, might involve mitochondrial damage as part of pathogenesis of cirrhosis. Hence, the choice of cryptogenic cirrhosis, as it would provide better insights into the role of UPRMT exclusive to cirrhosis and not confounded by other risk factors. Accordingly, we hypothesized a role of deregulated UPRMT and hepatocyte senescence in synergistically contributing toward the pathogenesis of cryptogenic liver disease.
Briefly, the work revealed accumulation of senescent hepatocytes in decompensated cirrhosis and compromised UPRMT as a key senescence-associated feature. Intriguingly, a strong UPRMT in compensated cirrhosis indicated its possible role in survival. This work also highlights the role of mitochondrial protease, Caseinolytic mitochondrial matrix peptidase proteolytic subunit (CLPP), which is a key player of UPRMT in preventing stress-induced premature senescence at least in cell culture system.
Results
Low Dose of Dox Induces Permanent Growth Arrest Similar to Senescence in Hepatoma Cells
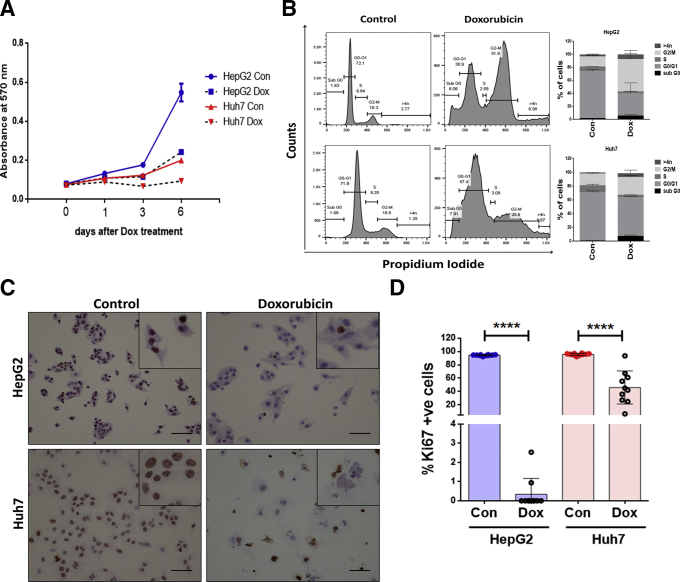
In a previous work we had demonstrated that low dose of Dox-induced senescence in osteosarcoma cells.20 To test if hepatoma cells (HepG2 and Huh7) can also show senescence-like changes, cells were treated with Dox for 2 hour with different doses ranging from 0.5 to 5 μM, followed by change into fresh medium and growth was monitored for 6 days. A 2 μM dose of Dox showed maximum growth arrest by sixth day in both the cell lines (Figure 1A). Flow cytometry analysis showed that Dox treatment resulted in significant growth arrest as revealed by 2–3-fold increase in percentage of cells in the G2/M phase of cell cycle in hepatoma cells. In comparison to G2M arrest, the percentage cell death as indicated by sub-G0 cells was considerably less in Dox-treated cells (Figure 1B). To test if the Dox-treated cells were permanently growth arrested, cells were stimulated with 10% serum and immunocytochemistry was performed for proliferation marker, Ki67. Compared with control, the Dox-treated cells on sixth day showed fewer number of Ki67-positive nuclei indicating growth arrest (Figure 1C and D). HepG2 cells with wild type p53, showed more prominent growth arrest compared with Huh7 cells harboring mutant p53.
Figure 1.
Dox induces growth arrest in hepatoma cells. (A) Growth curve of hepatoma cells (Huh7 and HepG2) treated with 2 μM Dox for 2 hours (day 0), replenished with fresh medium and growth kinetics was monitored at specified time intervals (1–6 days) by staining with crystal violet dye, whose absorbance was checked spectrophotometrically at 570 nm. Cells treated with vehicle alone served as Control (Con). (B) Cell cycle distribution of Control and Dox-treated cells as analyzed by flow cytometry. Following treatment with Dox for 2 hours, cells were cultured for 6 days, fixed in ethanol, stained with propidium iodide (PI), and subjected to flow cytometry for cell cycle analysis. Representative histogram and bar graph shows the distribution of cells in different phase of cell cycle. The data shown are mean ± SD from 3 separate experiments. (C) Immunocytochemistry for Ki67 in HepG2 and Huh7 control and Dox-treated cells after 6 days. Serum stimulation was done for 24 hours before Ki67 staining. Inset shows magnified view of cells. Brown color indicates Ki67-positive nuclei. Scale bar = 50 μm. (D) Bar diagram showing percentage Ki67 positivity in Control and Dox-treated hepatoma cells. Individual data points represent percentage positivity of at least 10 different microscopic fields and data are represented as mean ± SD. Student’s t test was used to calculate the significance. ****P ≤ .0001.
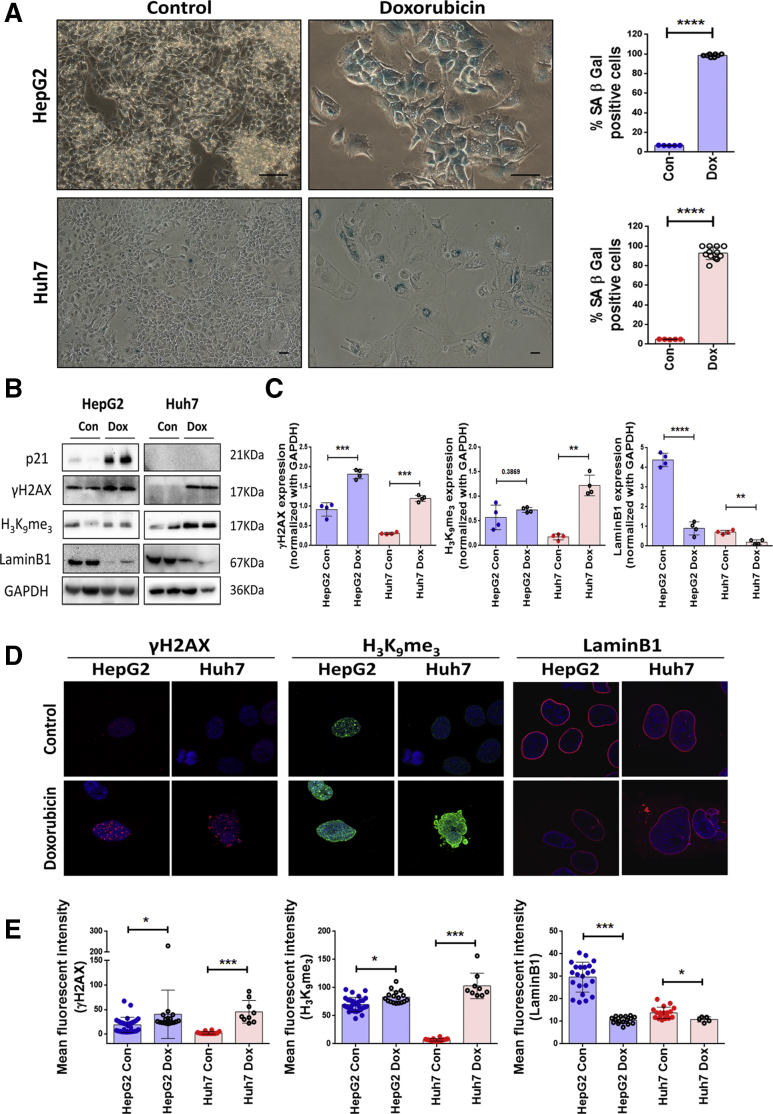
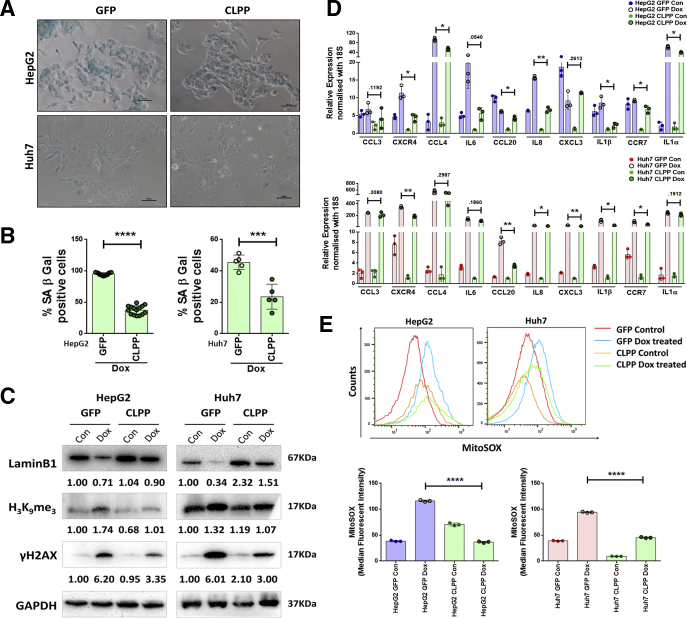
Dox-treated HepG2 and Huh7 cells under bright field microscope showed enlarged and flattened morphology and a significant increase in senescence-associated β-galactosidase (SA-β-gal) positivity (>90%) on the sixth day of treatment indicative of premature senescence (Figure 2A). Other senescence-associated markers were evaluated by both immunoblot assay and confocal microscopy. Premature senescence in Dox-treated cells was associated with increase in levels of DNA damage marker γH2AX and also a heterochromatin-associated repressive marker, H3K9me3 (Figure 2B–D). The inhibitor of cyclin-dependent kinases, p21, which results in growth arrest was prominently increased in the Dox-treated HepG2 cells. As p21 is a transcriptional target of p53, its levels were hardly detected in Huh7 cells harboring dysfunctional p53 (Figure 2B). The changes in these markers were also associated with a prominent loss in nuclear lamina protein, LaminB1, which was confirmed by immunoblotting as well as imaging (Figure 2B–D). Dox-treated HepG2 and Huh7 cells also showed a distinct senescence-associated secretory phenotype, which is described in detail subsequently. These results indicated that treatment with low dose of Dox-induced senescent features in hepatoma cells regardless of their p53 status. Hence, hereafter the Dox-treated cells are referred as senescent cells.
Figure 2.
Dox induces premature senescence in both HepG2 and Huh7 irrespective of their different p53 status. (A) Bright field microscopy images of Control and Dox-treated hepatoma cells (HepG2 with wild type p53 and Huh7 with mutant p53) subjected to SA-β-gal assay. Briefly, cells were treated for 2 hours with Dox, changed with fresh medium, and cultured for 6 days following which SA-β-gal assay was done. Blue color indicates SA-β-gal–positive cells. Scale bar = 50 μm. Adjoining bar diagram shows percentage SA-β-gal positivity in control and Dox-treated cells. Individual data points represent mean percentage positivity of at least 5 different microscopic view fields. The data are represented mean ± SD of multiple replicates. (B) Representative immunoblots showing expression of senescence-associated markers viz. p21, γH2AX, H3K9me3, and LaminB1 in Control and Dox-treated cells at sixth day. (C) Bar diagrams showing the relative expression of senescence markers in control and Dox-treated cells normalized with glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The experiment was done in quadruplicates and data were plotted as mean ± SD. (D) Merged confocal images showing expression of γH2AX (red), H3K9me3 (green), and LaminB1 (red) in control and Dox-treated cells on sixth day. Cells were counterstained with DAPI (blue). (E) Bar graphs show the relative mean fluorescent intensity (MFI) of senescence-associated markers calculated in different view fields. Individual data points represent MFI quantification in 1 particular field. Data are represented as mean ± SD. For all the experiments described, the significance was calculated by Student’s t test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001.
Dox-Induced Premature Senescence Is Associated With Mitochondrial Dysfunction and Compromised UPRMT
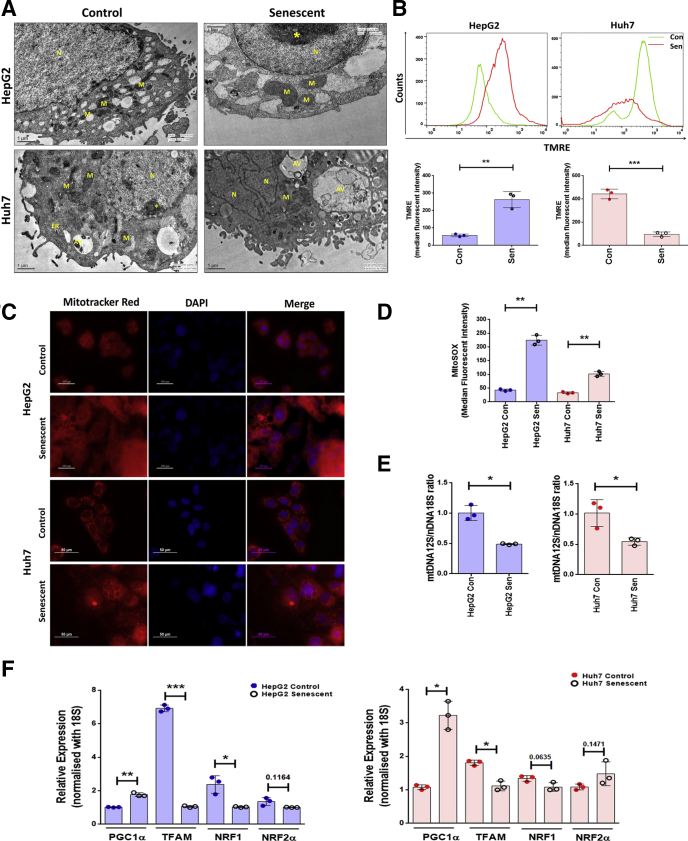
Transmission electron microscopy revealed fewer and enlarged mitochondria in senescent HepG2 and Huh7 cells (Figure 3A). Unlike the control cells which showed well placed stacks of mitochondrial cristae, the senescent cells showed incomplete and disrupted cristae. Mitochondrial membrane potential was measured using a mitochondrial membrane potential sensitive dye, tetramethylrhodamine ethyl ester (TMRE). Senescent HepG2 cells showed more accumulation of TMRE than control cells suggestive of hyperpolarized mitochondria (Figure 3B). In contrast, the Huh7 cells showed 4-fold decrease in TMRE accumulation, indicating mitochondrial depolarization. The TMRE results were also corroborated by yet another mitochondrial membrane potential–sensitive agent, Mitotracker red, which showed enhanced intensity in senescent HepG2 cells and appeared feeble in senescent Huh7 cells (Figure 3C).
Figure 3.
Mitochondrial dysfunction following senescence in HepG2 and Huh7 cells. (A) Representative transmission electron micrographs of control and senescent hepatoma cells. Scale bar = 1 μm. (B) Representative histogram plots showing changes in mitochondrial polarization as measured by sequestration of TMRE in Control and senescent (Sen) cells on sixth day as analyzed by flow cytometry. Bar diagrams shows quantification of TMRE in control and senescent hepatoma cells using 3 biological replicates. (C) Representative images of control and senescent cells stained with mitochondrial potential dependent dye and Mitotracker red (mitochondria, red) and counterstained with DAPI (nucleus, blue). Scale bars = 100 μm and 50 μm. (D) Bar diagram showing quantification of mitochondrial ROS production in control and senescent cells stained with fluorescent dye, MitoSOX, which is a probe to detect mitochondrial superoxide. (E) mtDNA quantification in HepG2 and Huh7 cells by real-time qPCR amplification of mitochondrial encoded 12S rRNA and the nuclear encoded 18S rRNA, which served as a reference gene. Results are expressed as relative ratio of 12S mtDNA over 18S nDNA. (F) Relative gene expression, by qPCR, of mitochondrial biogenesis regulatory genes in control vs senescent HepG2 and Huh7 cells. For all experiments the bar indicates values as mean ± SD of 3 independent experiments. P values were calculated by paired Student’s t test. *P ≤ .05, **P ≤ .01, ***P ≤ .001. The asterisk indicates heterochromatin. AV, autophagic vacuole; ER, endoplasmic reticulum; M, mitochondria; N, nucleus.
As oxidative stress is an underlying cause of cellular senescence we stained the cells with MitoSOX, a specific indicator of mitochondrial superoxide. Senescent HepG2 and Huh7 cells showed increase in levels of MitoSOX, as analyzed by flow cytometry (Figure 3D). Next, we evaluated if mitochondrial dysfunction is related to mitochondrial content. For this the mitochondrial DNA (mtDNA) content relative to nuclear DNA (nDNA) content (mtDNA/nDNA) was measured by real-time quantitative polymerase chain reaction (qPCR). The senescent cells showed a significant decrease in mtDNA (Figure 3E). As mitochondrial biogenesis is also responsible for the mitochondrial mass, we evaluated the expression levels of various genes involved in its biogenesis. Peroxisome proliferator-activated receptor gamma coactivator-1 alpha showed increased expression following senescence, while its downstream targets such as targets mitochondrial transcription factor A, nuclear respiratory factor 1, and nuclear respiratory factor 2 alpha showed downregulation (Figure 3F).
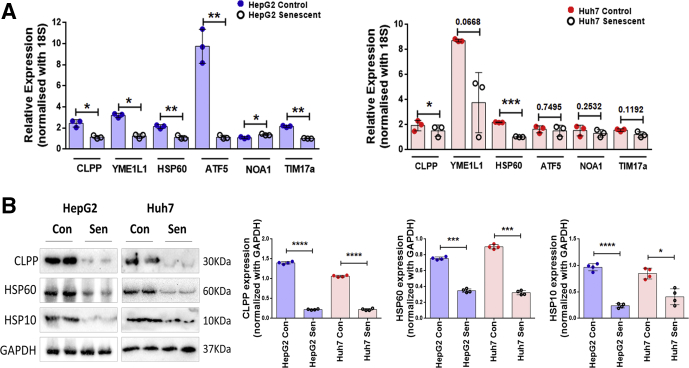
In the model organism, C. elegans, it has been shown that defects in mitochondria lead to activation of a stress response pathway (ie, the UPRMT). Some key players of UPRMT pathway were therefore analyzed in senescent cells by both real-time PCR and immunoblotting. Senescent HepG2 and Huh7 cells showed a significant decline in transcript levels of mitochondrial matrix proteases CLPP and YME1L1, mitochondrial chaperone HSP60, and a transcription factor (ATF5) (Figure 4A). Further immunoblotting revealed 2 fold reductions in key players of UPRMT pathway (ie, HSP60, HSP10, and CLPP) (Figure 4B), thereby indicating compromised UPRMT response during senescence.
Figure 4.
UPRMTis compromised in Dox-induced senescent cells. (A) Bar graphs represent relative expression of UPRMT genes as measured by real-time qPCR in control vs senescent HepG2 and Huh7 cells. The bars represent mean ± SD from 3 independent experiments. (B) Representative immunoblots of UPRMT genes in control and senescent HepG2 and Huh7 cells. The adjoining bar graph shows their relative quantification normalized to GAPDH. Data are represented as mean ± SD of 4 biological replicates. P values were calculated by paired Student’s t test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001.
Cryptogenic Cirrhotic Liver Showed Accumulation of Senescent Hepatocytes and an Altered UPRMT
Having identified the senescence-associated markers in the in vitro study, we evaluated if the these markers can also identify senescent hepatocyte in the end stage liver disease. In addition, the gene expression of the UPRMT pathway was also evaluated in the chronology of events leading to cirrhosis. Extrinsic factors such as alcohol, lipid, and hepatotropic viruses are known to compromise mitochondrial functions. Hence, these etiologies were excluded and the focus of the study was on cryptogenic liver disease in which there is no information available, to the best of our knowledge, on the molecular events involved in disease progression. A detailed workout on the different subject groups and their clinical and pathological characterization is described in the Material and Methods section. Briefly, the 4 groups included in the study are control subjects, fibrosis, and compensated and decompensated cirrhosis. The clinical and immunohistochemical parameters of the various subjects are shown in Table 1. As expected the decompensated liver showed a significantly higher Model for End-Stage Liver Disease (MELD) score, lowered levels of albumin, and deranged liver function tests.
Table 1.
Clinical Parameters and Immunohistochemical Scores of Various Markers in Different Patient Groups
| Variables | Control | Fibrosis | Compensated cirrhosis | Decompensated cirrhosis | P value (1-Way ANOVA) |
|---|---|---|---|---|---|
| Age, y | 26.20 ± 0.58 | 34.00 ± 5.77 | 55.75 ± 1.81 | 45.57 ± 3.85 | .0017a |
| Sex, % | 0.60 | 0.33 | 0.50 | 0.29 | |
| Total bilirubin, mg/dL | 0.88 ± 0.15 | 0.88 ± 0.16 | 1.73 ± 0.33 | 5.09 ± 1.91 | .0013a |
| AST, IU/L | 21.4 ± 3.03 | 55.50 ± 9.01 | 51.63 ± 6.81 | 60.43 ± 11.52 | .0111a |
| ALT, IU/L | 20.6 ± 6.64 | 88.83 ± 23.29 | 37.00 ± 6.50 | 31.43 ± 6.90 | .0061a |
| Albumin, g/dL | 4.26 ± 0.19 | 4.02 ± 0.25 | 3.58 ± 0.18 | 2.83 ± 0.25 | .0046a |
| Creatinine, mg/dL | 0.64 ± 0.07 | 1.06 ± 0.44 | 0.64 ± 0.04 | 0.68 ± 0.10 | .9903 |
| INR | 0.99 ± 0.03 | 1.02 ± 0.05 | 1.18 ± 0.08 | 1.81 ± 0.23 | .0015a |
| MELD score | 6.60 ± 0.40 | 8.67 ± 2.27 | 10.00 ± 0.89 | 17.29 ± 2.66 | .0035a |
| p21 (IHC score) | 3.70 ± 0.43 | 27.59 ± 3.94 | 55.79 ± 7.31 | 102.4 ± 7.24 | <.0001a |
| p53, % positivity | 34.73 ± 1.98 | 37.27 ± 1.29 | 58.80 ± 1.01 | 69.53 ± 1.56 | <.0001a |
| LaminB1, % intact nuclear membrane | 86.50 ± 0.92 | 86.70 ± 0.99 | 42.14 ± 1.55 | 6.81 ± 0.46 | <.0001a |
| γH2AX (IHC score) | 53.13 ± 2.72 | 80.00 ± 6.44 | 133.5 ± 3.16 | 239.6 ± 4.87 | <.0001a |
| ClpP (intensity score) | 1.00 ± 0.0 | 0.33 ± 0.21 | 2.94 ± 0.63 | 1.71 ± 0.29 | .0001a |
| HSP60 (intensity score) | 2.20 ± 0.37 | 1.83 ± 0.17 | 2.75 ± 0.25 | 2.00 ± 0.22 | .0390a |
| HSP10 (intensity score) | 2.00 ± 0.32 | 1.67 ± 0.21 | 2.75 ± 0.16 | 1.29 ± 0.18 | .0024a |
Values are mean ± SD unless otherwise indicated.
ALT, alanine aminotransferase; ANOVA, analysis of variance; AST, aspartate aminotransferase; IHC, immunohistochemistry; INR, international normalized ratio; MELD, Model for End-Stage Liver Disease.
Significant change with a level of significance of .05.
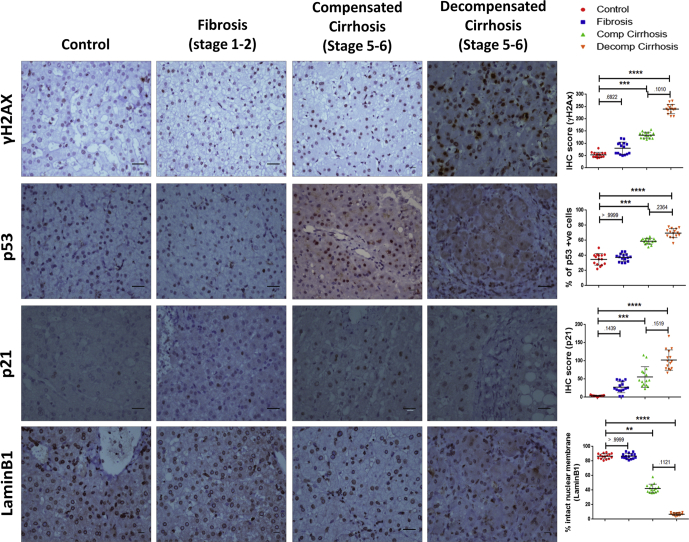
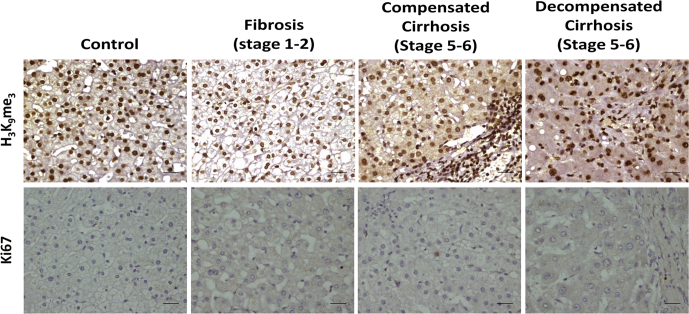
The senescence-associated gene signature (p21, p53, γH2AX, LaminB1, and H3K9Me3), was used to identify hepatocyte senescence in liver disease by performing immunohistochemistry on serial sections of 15 cases in each subject group of different histological stages ranging from fibrosis (stage 1–2) to cirrhosis (stage 5–6 with compensated and decompensated states). A progressive increase in staining of γH2AX in hepatocyte nuclei was noted in cirrhosis as compared with fibrosis indicative of persistent DNA damage (Figure 5). Accumulation of p53 and p21 in cirrhotic liver, together with loss of Ki67 (Figure 6) staining indicated growth arrest of hepatocyte. Expression of LaminB1 appeared solely on the nuclear membrane of hepatocytes in control and early fibrotic stage. Compensated cirrhotic liver showed a mixed pattern for LaminB1 expression, unchanged or with partial loss from nuclear membrane. However, LaminB1 appeared feebly stained and predominantly in nucleoplasm with either a complete or partial loss from the nuclear membrane in decompensated cirrhotic liver. Surprisingly, the staining intensity or level of H3K9me3 remained unaffected in the various stages of liver disease (Figure 6). These results indicated hepatocyte senescence as a key feature of cryptogenic liver disease, and loss of LaminB1 appeared to be a promising senescence-associated marker.
Figure 5.
Accumulation of senescent hepatocytes in various stages of liver disease. IHC showing senescence-associated markers in different subject groups (ie, Control, fibrosis, compensated [Comp] cirrhosis, and decompensated [Decomp] cirrhosis). A total of 15 different samples were analyzed in each group. Representative images show expression of DNA damage-associated marker γH2AX; growth arrest marker p53; cyclin-dependent kinase inhibitor p21, and nuclear lamina protein LaminB1. Brown staining indicates positivity. Scale bar = 10 μm. Adjacent scatter plots show the changes in expression of each of the markers in different stages of liver disease. For γH2AX and p21, IHC score was calculated as explained in Materials and Methods section. For p53, percentage positivity was plotted, whereas for LaminB1, percentage of cells with intact nuclear membrane was taken as variable for quantification. Individual data points in the scatter plot represent samples scored in each group. The quantified data are represented as mean ± SD. Kruskal Wallis 1-way analysis of variance with Dunn’s multiple comparison test was used to determine the statistical significance between the groups. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001.
Figure 6.
Representative images of H3K9me3and Ki67 immunohistochemistry done in various subject groups (ie, control, fibrosis, and compensated and decompensated cirrhosis). Brown color indicated positivity. Scale bar = 20 μm.
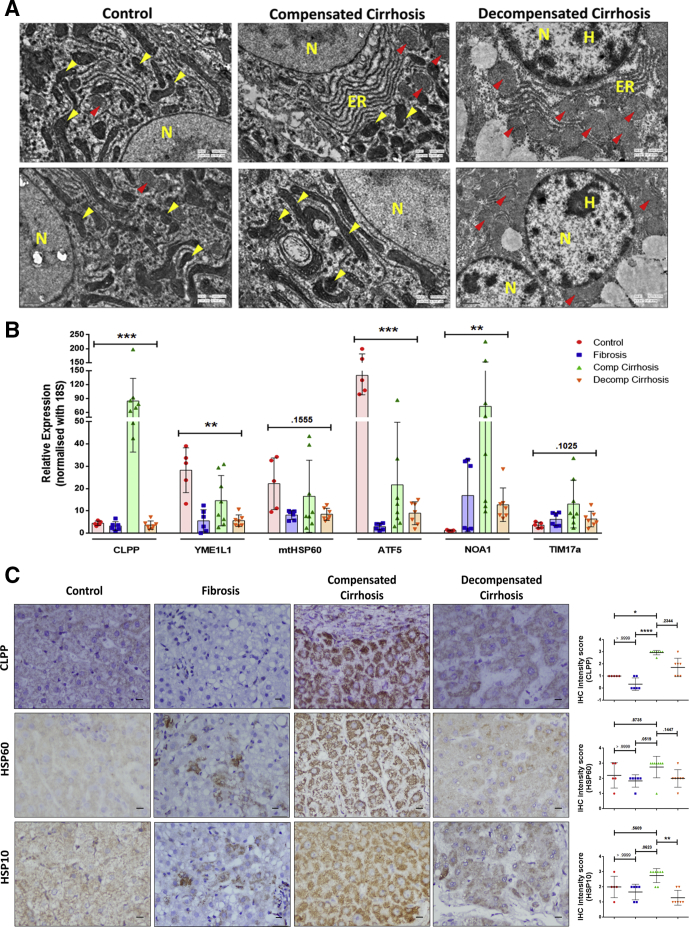
Next, we tested if progression toward decompensation is also associated with mitochondrial dysfunction. Transmission electron microscopy of control liver revealed presence of multiple intact electron dense mitochondria with well-formed cristae (Figure 7A). The compensated cirrhotic liver showed mitochondria with either well defined or poorly formed cristae. The decompensated cirrhotic tissue showed enlarged mitochondria with a loss of cristae. The alterations in mitochondrial morphology in liver disease were also associated with alterations in UPRMT pathway. The expressions of most UPRMT genes (CLPP, HSP60, TIM17, and NOA1) were relatively similar in control and fibrosis group. The compensated cirrhotic cases showed relatively higher RNA expression of UPRMT genes (CLPP, YME1L1, HSP60, and ATF5) when compared with decompensated cirrhosis and fibrosis (Figure 7B). Of these, mitochondrial protease, CLPP, showed the most significant increase in its transcript level in compensated cirrhosis. In immunohistochemistry also, an intense CLPP expression was noted in the compensated cirrhotic tissue, which also appeared punctate possibly indicative of its mitochondrial localization (Figure 7C). The other UPRMT markers (ie, HSP60 and HSP10) also showed an increase in immunohistochemistry (IHC) intensity scores in compensated cirrhosis compared with both control and decompensated state (Figure 7C). These results indicated a probable role of an activated mitochondrial stress response pathway as an adaptive response only in the compensated cirrhotic liver.
Figure 7.
Mitochondrial-associated changes in various stages of end-stage liver disease. (A) Representative transmission electron micrograph of liver tissue from control and compensated and decompensated cirrhosis. Note the presence of intact electron dense mitochondrial cristae (indicated by yellow arrowheads) in control and compensated cirrhosis, while enlarged mitochondria with diffused cristae (indicated by red arrowheads) in decompensated cirrhosis. Scale bar = 0.5 μm. (B) Bar graph shows the relative expression of UPRMT genes quantified by qPCR in control, fibrosis, compensated (Comp), and decompensated (Decomp) cirrhosis. Values are represented as mean ± SD. Individual data points represent number of subjects in each group: n = 5 in control, n = 6 in fibrosis, n = 8 in compensated cirrhosis, and n = 7 in decompensated cirrhosis. Kruskal-Wallis 1-way analysis of variance was used to calculate the significance within the groups. (C) Representative images of liver sections showing immunohistochemistry for UPRMT markers in control, fibrosis, compensated (Comp), and decompensated (Decomp) cirrhosis. Scale bar = 10 μm. Adjacent scatter plots representing variations in intensity scores (0 =absent, 1 = minimal, 2 = moderate, 3 = severe) of CLPP, HSP60, and HSP10. Data are represented as mean ± SD. Kruskal Wallis 1-way analysis of variance with Dunn’s multiple comparison test was used to calculate the significance between the groups. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001.
CLPP Overexpression in Hepatoma Cells Attenuates Senescence by Inhibiting ROS
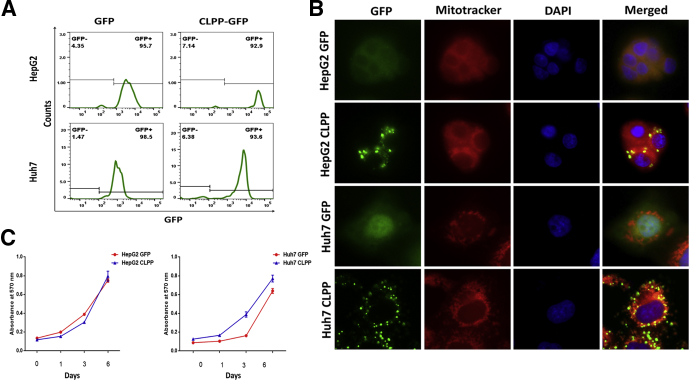
Amongst all the UPRMT genes CLPP showed maximal expression in compensated liver, hence it was overexpressed in hepatoma cells to evaluate its role in premature senescence. Both HepG2 and Huh7 cells were transfected with either CLPP-green fluorescent protein (GFP) or GFP alone and stable cell clones showing more than 90% GFP positivity were selected (Figure 8A). Unlike the control cells which showed a pan-GFP expression, the CLPP overexpressing cells showed a punctate pattern in the cytoplasm which co-localized with Mitotracker, thereby confirming its mitochondrial localization (Figure 8B). The growth kinetics of CLPP overexpressing cells was similar to their control counterpart (Figure 8C). When treated with Dox, the CLPP-GFP cells showed almost 50% reduction in SA-β-gal staining when compared with control GFP cells (Figure 9A and B). This was also accompanied with increased levels of LaminB1 and decreased levels of γH2AX and H3K9Me3 in Dox-treated CLPP cells (Figure 9C). Senescence is often accompanied by a secretory phenotype, which was much subdued in CLPP expressing cells when exposed to Dox (Figure 9D).
Figure 8.
CLPP overexpression and establishment of stable hepatoma cell lines. (A) Histogram plots showing percentage of GFP positivity in stable HepG2 and Huh7 cells expressing either empty GFP vector or CLPP-GFP as analyzed by flow cytometry. (B) Immunofluorescence images of stable cell clones of HepG2 and Huh7 cells showing expression of either GFP or CLPP-CLPP (green). Note that CLPP-GFP showed a punctate expression which co-localized with Mitotracker red. Nucleus was counterstained with DAPI. (C) Growth of cells expressing GFP and CLPP-GFP was monitored by uptake of crystal violet dye at specified time intervals and quantified by reading the absorbance at 570 nm. The data are represented as mean ± SD from 3 independent experiments.
Figure 9.
CLPP overexpression attenuated senescence-associated changes in hepatoma cells following Dox treatment by reducing ROS levels. (A) Representative bright field microscopic images of SA-β-gal staining in GFP vector and CLPP-GFP overexpressing HepG2 and Huh7 cells following Dox treatment on sixth day. Scale bar = 50 μm. (B) Bar diagrams showing percentage of SA–β-gal–positive cells. Individual data points represent percentage positivity per microscopic field. (C) Immunoblots showing the expression of 3 senescence-associated markers and GAPDH served as loading control. The numbers below the blots represent relative fold change compared with untreated Control in GFP and CLPP overexpressing cells following Dox treatment at sixth day. (D) Relative expression of senescence-associated secretory phenotype markers as quantified by real-time qPCR in Control and Dox-treated HepG2 and Huh7 cells expressing either GFP or CLPP-GFP. The data represent mean ± SD from 3 independent experiments. (E) Representative histogram showing shift in MFI of MitoSOX, which is an indicator of the mitochondrial ROS levels. The bar diagram showing quantification of mitochondrial ROS in HepG2 and Huh7 cells expressing either GFP or CLPP-GFP when treated with either vehicle or Dox. MFI of MitoSOX was calculated on GFP+ gated population. Individual data points represent 3 biological replicates and data are represented as mean ± SD. For all statistical significance, P values were calculated by Student’s t test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001.
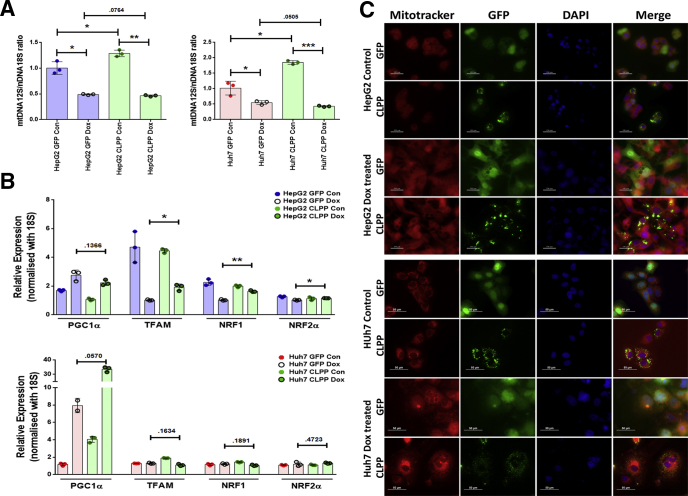
Next, we examined the probable mechanisms by which CLPP could attenuate cellular senescence. Several studies have implicated ROS in regulating cellular senescence. To examine the mitochondrial-specific ROS generation, cells were stained with MitoSOX and analyzed by flow cytometry. The Dox-treated senescent HepG2 and Huh7 cells expressing GFP alone showed higher ROS levels on sixth day compared with nonsenescent counterparts (Figure 9E). However, the CLPP overexpressing cells showed a significant decline (3-fold in HepG2 and 2-fold in Huh7) in the ROS levels after Dox treatment on sixth day (Figure 9E). Besides ROS level, we also evaluated if CLPP can alter mitochondrial functioning by effecting either its number or biogenesis. Ratio of the mtDNA (12S)/nDNA (18S) gene was used to quantify changes in mtDNA levels. Expression of CLPP marginally enhanced 12S/18S ratio in absence of Dox stress. Intriguingly, following Dox treatment both vector alone and CLPP overexpressing cells showed similar low level of mitochondrial DNA (Figure 10A). This indicated that CLPP is not able to increase the mitochondrial DNA content following stress. Multiple transcription factors play a role in mitochondrial biogenesis, of which main regulators are peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) and its downstream targets mitochondrial transcription factor A (TFAM) and nuclear respiratory factors (NRF) 1 and 2. CLPP on its own was unable to alter the expression levels of regulators involved in the mitochondrial biogenesis pathway in absence or presence of Dox-induced stress (Figure 10B). Because mitochondrial biogenesis remained unaffected by CLPP, we next tested the possibility if CLPP can rescue the damaged mitochondria by restoring their polarization state. Overexpression of CLPP could not change the polarization status of mitochondria in both the cell lines as evident by Mitotracker staining (Figure 10C). These results thus indicate that CLPP has little role in mitochondrial biogenesis and polarization status in conditions of stress-induced premature senescence. Overall the results indicated that CLPP overexpression can reduce senescence by lowering the oxidative stress and the resulting DNA damage.
Figure 10.
Effect of CLPP on mitochondrial DNA content, biogenesis, and polarization in HepG2 and Huh7 cells. (A) mtDNA quantification in hepatoma cells expressing either GFP or CLPP and treated without (Con) or with Dox. Quantification was done by real-time qPCR amplification of mitochondrial 12S rRNA gene and nuclear 18S rRNA, which served as a reference gene. Results are expressed as ratio of 12S mtDNA to 18S nDNA. (B) Relative gene expression of mitochondrial biogenesis regulatory genes by real-time qPCR in GFP vs CLPP overexpressing cells treated with Dox. Untreated cells served as control. The bar diagram represent values plotted as mean ± SD from 3 different experiments. P values were calculated by paired Student’s t test. *P ≤ .05, **P ≤ .01, ***P ≤ .001. (C) Representative fluorescence images of control and Dox-treated GFP and CLPP-GFP (green) overexpressing cells, stained with Mitotracker (mitochondria, red) dye, which detects mitochondrial polarization status. Cells were counterstained with DAPI (nucleus, blue). Scale bars = 100 μm and 50 μm.
CLPP Overexpression Rescues Senescent Phenotype in Non-Neoplastic Hepatocytes, PH5CH8
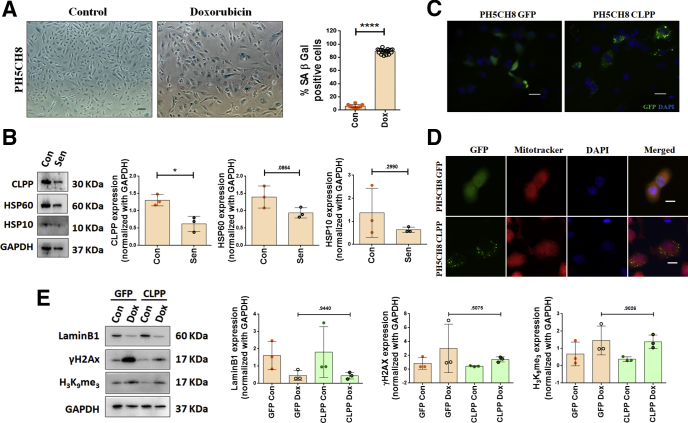
As HepG2 and Huh7 are hepatoma cells, we repeated some of the experiments in PH5CH8 cells, which are non-neoplastic immortalized human hepatocytes and closely resembles the primary hepatocytes.21 Dox treatment led to induction of senescence in PH5CH8 cells with almost 90% cells showing SA-β-gal positivity (Figure 11A). The senescent PH5CH8 cells also showed lower levels of UPRMT players (e, CLPP, HSP60, and HSP10), with a significant 2-fold reduction in CLPP expression in the senescent cells when compared with the control (Figure 11B). Hence, CLPP was transiently transfected into PH5CH8 cells. CLPP-GFP showed a distinct punctate pattern in the cytoplasm, unlike that of pan-localization pattern of GFP vector alone (Figure 11C). Further, CLPP-GFP colocalized with Mitotracker red, confirming its mitochondrial localization in the nontumorigenic hepatocyte cell line (Figure 11D). On the sixth day following treatment with Dox, the CLPP-overexpressing PH5CH8 showed an increase in levels of LaminB1 and decrease in levels of γH2AX and H3K9Me3 compared with the control (Figure 11E). However, unlike the hepatoma cells, the changes in levels of these senescent markers in PH5CH8 were not statistically significant. This in turn is explained by the fact that PH5CH8 cells are difficult to transfect with only 30% transient transfection efficiency.
Figure 11.
CLPP overexpression rescues senescent phenotype in non-neoplastic hepatocytes (PH5CH8). (A) Representative bright field microscopy images of SA-β-gal staining in Control and Dox (2 μM)–treated cells. Cells were treated for 2 hours, changed to fresh medium, and grown for 6 days, following which staining was done. Scale bar = 50 μm. Adjoining bar diagram shows percentage SA-β-gal positivity with data points representing at least 10 different microscopic fields. (B) Representative immunoblots of UPRMT genes in Control and senescent (Sen) PH5CH8 cells. The adjoining bar graphs shows their relative quantification normalized to GAPDH. Individual data points represent 3 biological replicates. (C) Representative fluorescence images showing expression of GFP and CLPP-GFP (green) transiently transfected in PH5CH8 cells. Nucleus was counterstained with DAPI (blue). Scale bar = 20 μm. (D) Immunofluorescence images of PH5CH8 cells showing expression of either empty GFP vector or CLPP (green). As compared with pan expression of GFP vector control, CLPP showed a punctate expression which co-localized with Mitotracker Red. Nucleus was counterstained with DAPI (blue). Scale bar = 10 μm. (E) Representative immunoblots of senescence-associated markers in GFP and CLPP-GFP overexpressing PH5CH8 cells after Dox treatment. The adjoining bar graph shows the relative quantification normalized to GAPDH. Individual data points represent 3 biological replicates. The bar diagrams represent data values as mean ± SD. P values were calculated by Student’s t test. *P ≤ .05, ****P ≤ .0001.
Effect of CLPP on Mitochondrial Respiration
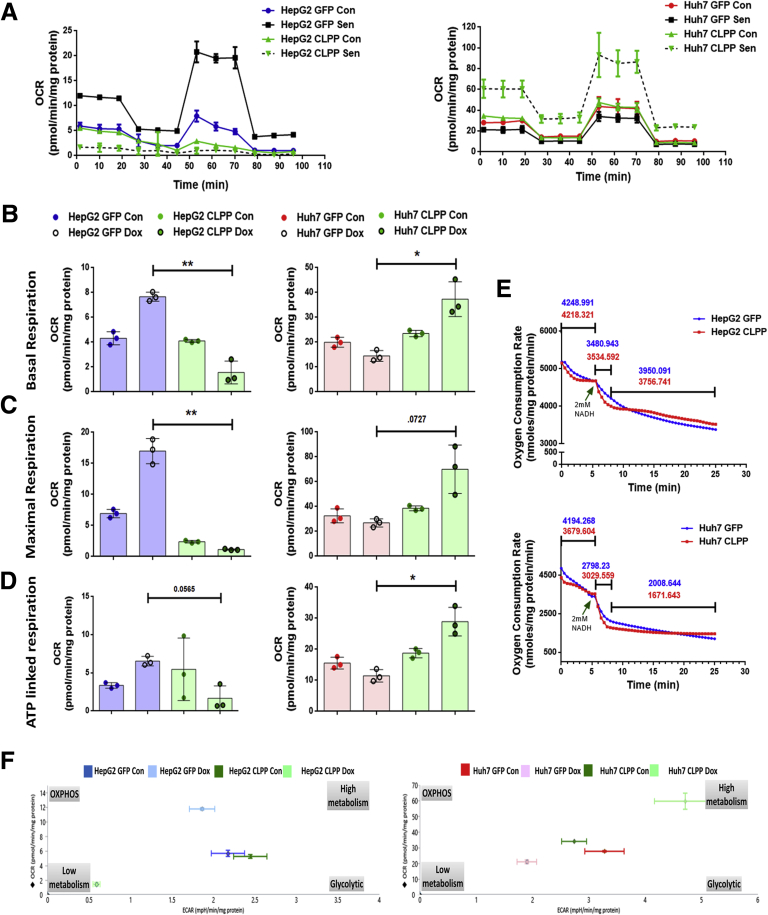
As CLPP is localized in mitochondria, an obvious question was whether it can affect the oxygen consumption and bioenergetics profile following Dox stress. The overall respiratory responses was measured using the Seahorse extracellular flux analyzer, for the hepatoma cells expressing either GFP vector or CLPP and additionally cells were treated with and without Dox. The respiratory response is illustrated in Figure 12A. In general, Huh7 cells exhibited higher basal rate of respiration compared with HepG2 cells (Figure 12A and B). Compared with control, the basal respiration rate was higher in senescent HepG2 cells, while Huh7 senescent cells showed a subtle, albeit nonsignificant, decrease. Similar to the extracellular flux assay, the respiratory capacity of isolated mitochondria using an oxygen microsensor (TBR1025: One-Channel Free Radical Analyzer), in presence of CLPP, was barely affected (Figure 12E). Overall, in the absence of Dox-induced stress, CLPP overexpression on its own did not alter the basal respiration profile in both the hepatoma cell lines (Figure 12A, B, and E). Following Dox treatment, HepG2-CLPP showed a decrease in basal, maximal, and adenosine triphosphate–linked respiration (Figure 12B–D) in contrast to the marked increase in treated Huh7-CLPP cells, compared with Dox-treated GFP cells (Figure 12B–D). To get an overall picture of the bioenergetics profile of CLPP overexpressing cells in presence and absence of Dox-induced stress, the basal oxygen consumption rate (OCR) vs extracellular acidification rate data was plotted (Figure 12F). The energy map showed HepG2 having a glycolytic phenotype, while Huh7 cells showed a high metabolic phenotype with higher glycolysis and oxidative phosphorylation (OXPHOS). On its own, CLPP shifted the bioenergetic profiles in presence of Dox stress, but not in its absence. Dox-treated HepG2-GFP senescent cells showed high metabolic state, while HepG2-CLPP Dox-treated cells showed a low glycolytic profile. Huh7-GFP Dox-treated senescent cells had glycolytic phenotype whereas, Huh7-CLPP Dox–treated cells showed higher energetic state of both OXPHOS and glycolysis. Overall, these results indicated that CLPP affects the mitochondrial respiration only when cells are exposed to stress conditions, and, further, it shifts the energy phenotype depending on the cellular context.
Figure 12.
CLPP overexpression altered mitochondrial respiration in hepatoma cells. (A) Representative extracellular flux assay plots of HepG2 and Huh7 cells, expressing either GFP or CLPP, using the Seahorse XF Cell Mito Stress Test Kit. Respiration was monitored on sixth day following treatment with either Dox or vehicle (Con). The OCR was monitored at basal level in the absence of exogenous substrate, followed by sequential addition of oligomycin (1.0 μM), FCCP (0.50 μM for HepG2 and 0.25 μM for Huh7), and rotenone/antimycin A (0.5 μM). Points represent mean ± SD. Protein content of each well was used to normalize the OCR. (B) Bar graphs represent basal respiration in both HepG2 and Huh7 cells expressing either GFP or CLPP. (C) Bar graphs represents maximal respiration in both HepG2 and Huh7 cells expressing either GFP or CLPP. (D) Bar graphs represent adenosine triphosphate linked respiration in both HepG2 and Huh7 cells expressing either GFP or CLPP. (E) OCR of isolated mitochondria from GFP and CLPP overexpressing HepG2 and Huh7 cells as measured by oxygen microsensor (TBR1025 single-channel free radical analyzer). 2 mM Nicotinamide adenine dinucleotide was added (arrow) after 5 minutes and its oxidation was monitored every 30 seconds. (F) Energy phenotype analysis by plotting OCR vs extracellular acidification rate in CLPP and GFP overexpressing cells with or without Dox treatment. The bar graph represents value as mean ± SD and P values was calculated by Student’s t test. *P ≤ .05, **P ≤ .001.
Discussion
Hepatocyte senescence resulting in growth inhibition is often a terminal event following liver injury. Strategies to prevent senescence are likely to stop end-stage liver disease progression. In fact, a recent study has suggested that regenerative capacity of liver can be improved by specifically targeting the senescent hepatocytes.22 The present study describes UPRMT as a key pathway to prevent senescence-associated dysfunction. Decline in UPRMT exaggerates the hepatocyte senescence and shifts the cirrhotic fate toward decompensation.
Hepatocyte Senescence Associated With Loss of LaminB1 Is a Prominent Feature of Decompensated Cryptogenic Cirrhosis
Unlike the molecular events associated with alcoholic, viral, and fatty liver disease etiologies, hardly any data are available on cryptogenic liver disease. The present study for the first time indicates accumulation of senescent hepatocytes in cryptogenic cirrhosis, which in turn was accompanied by persistent DNA damage (γH2AX), mitoinhibition (low Ki67 index and increase in cell cycle inhibitors p21 and p53), together with loss of nuclear lamina protein LaminB1. However, it is important to note that increased levels of p53 and p21 can also help maintain quiescence in hepatocytes23, 24 and hence more specific markers of senescence are required. We now report that loss of LaminB1 is a promising marker to detect senescent hepatocytes in liver disease. The nuclear lamins belong to class of intermediary filaments which help in maintaining the nuclear morphology, structure, and function.25 Recent studies have indicated mutations in lamin or lamin-associated genes in fatty liver disorders and knockout of lamin in rodent model resulted in spontaneous fatty liver development.26, 27 In view of a role of lamin in gastrointestinal and liver disease,28 we surmise that premature senescence-associated with loss of LaminB1 as a possible cause of cryptogenic cirrhosis.
Compensated Cirrhosis Mounts a Strong UPRMT, Which Is Compromised in Decompensated Cirrhotic Liver and Senescent Hepatocytes
The present data indicated a specific increase in UPRMT pathway in compensated cirrhosis compared with both early fibrosis and late decompensated state. The present study is important as the data is scanty regarding the molecular event(s) that help in preserving organ function during compensatory phase of cirrhosis. Using a rodent cirrhotic model Liu et al29 constructed different gene clusters in compensated vs decompensated liver. Of these, the gene cluster with nuclear factor-kappa B as a hub protein showed an initial increased expression during early cirrhosis followed by a decrease in later stages. Our results on UPRMT signaling is similar to the observations of Liu et al29 as compensated cirrhotic liver mounted a strong UPRMT signaling, which in contrast was attenuated in decompensated liver. In fact, the electron micrographs showed near-normal mitochondria in compensated cirrhosis, thereby indicating a possible role of UPRMT in maintaining organelle integrity. This probably could explain why even in presence of senescent cells, the compensated liver could retain its function. Thus, we infer that UPRMT is a compensatory mechanism mounted during early phase of cirrhosis to help maintain the hepatocyte function; however, beyond a certain limit, a precipitous decline in the mitochondrial stress pathway can lead to progressive phase of decompensation. While a role of hepatocyte senescence has been shown in liver disease due to alcohol, fatty liver, and viral etiologies,30 it remains to be seen if the role of UPRMT seen in cryptogenic cirrhosis is also applicable to other known causes of liver disease. Nonetheless, the present work indicated UPRMT as a promising therapeutic target in liver disease.
Overexpression of Mitochondrial Protease, CLPP Attenuates Senescence Response, Reduces Oxidative Stress, and Alters the Mitochondrial Respiration
The contrasting difference in UPRMT in compensated and decompensated cirrhotic liver led us to reason that UPRMT pathway may provide a survival advantage and even protect cells from stress-induced senescence. Indeed, CLPP overexpression suppressed senescence-associated markers in both the hepatoma cell lines regardless of their p53 status. Our results are also supported by a report that loss of CLPP accelerated replicative senescence in mouse fibroblasts.31 Additionally, knockdown of CLPP in muscle cells led to decrease in proliferation, which was not due to cell death.32 There are contradictory results on CLPP knockout and ageing. Gispert et al31 reported that CLPP knockout mice are smaller sized and have high postnatal mortality. On the contrary, yet another mouse model showed that loss of CLPP resulted in death in utero and those pups that survived showed normal aging.33 The probable mechanism by which CLPP attenuates senescence response is mostly by lowering the levels of reactive oxygen species. The mitochondrial protease complex of LON-CLPP can degrade the mitochondrial complex I ROS generating domain, thereby reducing ROS levels of the depolarized mitochondria.34 Also, knocking out CLPP in cancerous cells resulted in increased ROS levels.35 As oxidative stress is a major initiator of senescence response, restoring mitochondrial protease activity may prove beneficial as an antioxidative strategy. As CLPP and other UPRMT markers are upregulated in compensated liver cirrhosis, we surmise that mitochondrial retrograde response may act as an effective strategy in maintaining mitohormesis during liver injury.
We found that CLPP influenced respiration only under conditions of oxidative stress. Intriguingly, in the 2 cell lines tested, CLPP exerted totally opposing effects on respiration; a decrease in OCR was seen in HepG2 cells and a contrary increase was observed in Huh7 cells. In view of these confounding results, we surmise that an effect of CLPP on respiration to prevent senescence is dependent on the cellular context. Knockdown of CLPP either in muscle cell line C2C12 or in leukemic cells resulted in lower basal respiration rate,32, 35 but loss of CLPP resulted in increase in basal respiration in white adipose tissue.18 Following CLPP knockout, only a moderate respiratory deficiency was reported in heart cells.36 Thus, it is inferred that effect of CLPP on respiration is dictated by not only the cellular context, but also the stress incurred. The present study, notwithstanding the small number of clinical samples, highlights a role of UPRMT as a potential drug target in liver disease.
Cryptogenic liver disease is generally believed to be burnt-out cases of autoimmune hepatitis (AIH), occult alcoholism and fatty liver, however, a recent study by Thuluvath et al,37 indicated that cryptogenic disease is a distinct entity. Whatever may be the initial cause, the subsequent molecular events in progression of cryptogenic disease remained elusive until now. In this regard, the present study is novel, as it has unraveled a role of hepatocyte senescence together with decline in UPRMT signaling as key events in development of cryptogenic liver disease. In conclusion, mounting either a strong UPRMT or restoring CLPP level appears to be a promising mitohormetic strategy in not only reducing cellular senescence by preventing oxidative stress, but also augmenting hepatocyte function to prevent cirrhosis-associated decompensation.
Materials and Methods
Cell Lines and Reagents
HepG2 (p53+/+) and Huh7 cells (p53mut) were obtained from National Centre for Cell Science (Pune, India). PH5CH8 cells were a kind gift from Professor Nobuyuki Kato (Okayama University, Japan). All cell lines were grown in Dulbecco’s modified Eagle medium (Cell Clone-Cat#CC3004) containing 10% fetal bovine serum (Hyclone-Cat#SH30071), 100 mg/mL penicillin, 100 mg/mL streptomycin, 2.5 μg/mL Amphotericin B (Hyclone-Cat#SV30079), and 2.5 mM L-glutamine at 37°C with 5% CO2 in a humidified incubator. Various antibodies with their dilutions and source of fluorescent dyes are given in Table 2.
Table 2.
List of Antibodies and Various Fluorescent Dyes Used in the Study
| Antibody | Catalog No. | Lot No. | Dilution |
||
|---|---|---|---|---|---|
| IHC/ICC | Immunofluorescence | Western blotting | |||
| p21 | AM434-5M (BioGenex) | AM2391115 | Ready to use | — | 1:500 |
| p53 | AM239-5M (BioGenex) | AM2391115 | Ready to use | — | — |
| Ki67 | AM297-5M (BioGenex) | Clone MIB-1 | Ready to use | — | — |
| H3K9me3 | ab8898 (Abcam) | GR186864-1 | 1:400 | 1:200 | 1:1000 |
| γH2AX | ab26350 (Abcam) | GR90011-1 | 1:500 | 1:200 | 1:1000 |
| GAPDH | G9545 (Sigma) | 060M4775 | — | — | 1:5000 |
| LaminB | sc-6216 (Santa Cruz Biotechnology) | F1812 | 1:100 | 1:50 | 1:500 |
| CLPP | GTX104656 (GeneTex) | 39708 | 1:200 | — | 1:500 |
| HSP10 | PAB501Hu01 (Cloud-Clone Corp) | A20170621181 | 1:20 | — | 1:250 |
| HSP60 | PAA822Hu01 (Cloud-Clone Corp) | A20170621167 | 1:20 | — | 1:250 |
| Alexa Fluor 488 or 594 conjugated secondary (anti-mouse/rabbit/goat) | anti-M-488 A11029 | 1503602 | — | 1:2000 | — |
| anti-M-594 A11032 | 1420905 | ||||
| anti-R-488 A11034 | 1423009 | ||||
| anti-R-594 A11037 | 1420978 | ||||
| Anti-Goat IgG-HRP | A5420 (Sigma-Aldrich) | whole molecule | — | — | 1:3000 |
| Anti-Rabbit IgG-HRP | 170-6515 (Bio-Rad) | whole molecule | — | — | 1:5000 |
| Anti-Mouse IgG-HRP | 170-6516 (Bio-Rad) | whole molecule | — | — | 1:5000 |
| Propidium iodide | 349523 (BD Biosciences) | ||||
| DAPI mounting media | H1200 (Vector Laboratories) | ||||
| MitoTracker Red CMXRos | M7512 (Invitrogen) | ||||
| MitoSOX | M36008 (Invitrogen) | ||||
| TMRE | 87917 (Sigma-Aldrich) | ||||
HRP, horseradish peroxidase; ICC, immunocytochemistry; IHC, immunohistochemistry; TMRE, tetramethylrhodamine ethyl ester perchlorate
Clinical Samples and Working Out of Subjects
The study included 4 different groups: healthy control, fibrosis, compensated cirrhosis, and decompensated cirrhosis, with 15 subjects in each group. Our hospital is a tertiary referral centre for liver diseases and in the prospective study done between 2015 and 2016, an average of about 733 patients with cirrhosis required admission of which 5.9% cases (43 of 733) were with unrecognized etiologies. These subjects tested negative for hepatitis B virus-DNA and hepatitis C virus-RNA. The sera of these subjects were also negative for autoantibodies, thereby ruling out autoimmune disorders. Further, these subjects had no genetic or family history of liver disease and did not have history of significant alcohol consumption. As most of the cryptogenic cases are believed to be burned-out nonalcoholic steatohepatitis, in the present study, patients with histological hepatic steatosis of <5% were also included. The biopsy of all the included patients had no evidence of any other possible etiology for liver cirrhosis.
The clinical staging of cirrhosis was classified into compensated or decompensated groups based on absence or presence of any evidence of decompensation—variceal bleed, hepatic encephalopathy, ascites, or jaundice.38 All the cases included in the present study were received for routine diagnostic reporting in the Department of Pathology. The pathological review of tissue samples and reassessment of certain histopathological features was performed by single pathologist.
Control liver tissue was obtained from farthest, nontumor area (>2 cm) of surgically resected specimens from patients undergoing surgery for cholangiocarcinoma and showing no specific pathology of liver. For fibrosis and compensated cirrhosis, liver biopsy was obtained from patients. Diagnosis of compensated group was based on the absence of clinical criteria of decompensation. Explant cirrhotic liver tissue was collected from patients undergoing live donor liver transplant surgery and this group mainly constituted the cryptogenic decompensated group.
Fibrosis group comprised cases showing fibrous expansion of some portal areas, with or without short fibrous septa and no evidence of bridging. Histological subclassification of cirrhosis was performed according to Laennec staging system, a modification of Metavir system.39
Immunohistochemistry for senescence-associated markers were done in all the subjects (n = 15 in each group). However, UPRMT expression was studied in a subset of subjects (control 5, fibrosis 6, compensated cirrhosis 8, and decompensated cirrhosis 7) in which enough material was available to do expressions by both qPCR and IHC. Samples were collected after approval from institutional ethics committee (approval # F25/5/43/AC/ILBS/2013/1157) according to Helsinki declaration and informed consent was taken from all the patients.
Treatment of Cells With Dox for Induction of Senescence
HepG2 and Huh7 were treated with 2 μM Dox (Cat#D1515; Sigma-Aldrich, St Louis, MO) for 2 hours in complete Dulbecco’s modified Eagle medium. However, for stable cells (expressing GFP or CLPP-GFP), a lower dose of 1 μM Dox was used. This is because a 1 μM dose was found to be sufficient for inducing senescence in both GFP control and CLPP-overexpressing cells. As a 2 μM dose of Dox also resulted in cell death in stable cells, to delineate effects specific to senescence, the lower dose of Dox was preferred. Cells were washed once and then replenished with media containing 10% fetal bovine serum. Medium was changed on third day. Induction of senescence on sixth day was confirmed by SA-β-gal assay as previously described by Dimri et al.40 Briefly, cells were fixed with 2% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline, and then washed with phosphate-buffered saline. Cells were stained for 3–24 hours in X-gal staining solution (1 mg/mL X-gal, 40 mM citric acid/sodium phosphate [pH 6.0], 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 150 mM NaCl, 2 mM MgCl2) at 37°C in a non-CO2 incubator. Cells stained with characteristic blue color were identified and counted using bright field microscopy.
Growth Assay by Crystal Violet
A total of 10,000 cells were plated in 24 well plates in triplicates and treated with Dox the following day. Cells were stained with 0.2% crystal violet made in 2% ethanol at indicated time intervals after Dox treatment and collected in 1% sodium dodecyl sulfate. The absorbance was recorded at 570 nm.
Cell Cycle Analysis by Flow Cytometry
Cells were fixed in 70% ethanol, washed, stained with propidium iodide solution containing 5 mg/mL RNase A at 37°C in the dark, and acquired in a flow cytometer (BD FACSCalibur; BD Biosciences, San Jose, CA) and DNA content was measured at 630/22 nm filter. Data analysis was done by FlowJo software Version 10.2 (Becton-Dickinson, Franklin Lakes, NJ).
Immunofluorescence Imaging
Cells plated on coverslips were fixed with either methanol/acetone (1:1) or 4% formaldehyde in phosphate-buffered saline. Blocking and permeabilization was done in 3% bovine serum albumin containing 0.1% Triton X-100 followed by incubation with primary antibody diluted in 1% bovine serum albumin overnight at 4°C. The primary antibody was detected using Alexa Fluor 488 or 594 conjugated secondary (anti-mouse/rabbit/goat) antibodies and mounted in DAPI mounting media. Images were taken either with confocal microscope (LEICA TCS SP2 Confocal Laser Scanning Microscope; Leica Microsystems, Heidelberg, Germany) or a fluorescence microscope (Nikon ECLIPSE Ni; Nikon Instruments Inc, Melville, NY). Mean fluorescent intensity of confocal images were calculated using LAS AF Lite software (Leica Microsystems).
Immunoblotting
Cells were lysed in RIPA lysis buffer containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mg/mL aprotinin) and phosphatase inhibitors (1 mM sodium orthovanadate, 10 mM sodium fluoride). Equal amount of protein was separated onto 12% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membrane. Blots were blocked in 5% nonfat dry milk, incubated with respective primary antibodies followed by incubation with horseradish peroxidase–conjugated secondary antibody. A single blot was cut and probed for different antibodies. The signals were detected by Clarity Max Western Enhanced Chemiluminescence substrate (Bio-Rad, Hercules, CA) using chemiluminescence detection system (Proteinsimple, San Jose, CA). Quantification of immunoblots was done using Image J software Version 1.51j8 (National Institutes of Health, Bethesda, MD).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
RNA was extracted from cell lines and tissue samples using TRIzol (Cat#15596018; Invitrogen, Carlsbad, CA) method according to manufacturer’s protocol. Complementary DNA (cDNA) was synthesized from 1 μg RNA using RevertAid First Strand cDNA Synthesis Kit (Cat#K1622; Thermo Fisher Scientific, Waltham, MA). qPCR was performed using Maxima SYBR Green (Cat#K0222; Thermo Scientific) in ABI ViiA 7 detection system (Applied Biosystems, Foster City, CA). Gene expression was normalized against 18S mRNA, which served as an internal control. Relative expression was calculated using the formula 2–ΔΔCt.41
Mitochondrial Polarization and Mitochondrial ROS Detection
For mitochondrial polarization, cells were trypsinized, washed with phosphate-buffered saline and stained with 100 nM TMRE in phosphate-buffered saline containing 1% fetal bovine serum for 20–30 minutes at 37°C in the dark. After incubation, cells were washed twice with staining buffer (phosphate-buffered saline, 1% fetal bovine serum) and acquired in BD FACSVerse (BD Biosciences, San Jose, CA). For imaging, cells were stained with 100 nM MitoTracker in staining buffer for 30 minutes at 37°C, washed and fixed in 4% formaldehyde for 15 minutes. Images were taken in a fluorescence microscope. For estimating mitochondrial ROS, cells were suspended in staining buffer containing 5 μM MitoSOX for 10 minutes at 37°C in the dark, washed and acquired in BD FACSVerse. Data were analyzed on GFP+ gated population using FlowJo software.
Transmission Electron Microscopy
A total of 4 × 106 cells (n = 3 for each sample) or 1 mm3 for tissue (2 specimens from each subject) were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde, made in 0.1M sodium phosphate buffer (pH 7.4) overnight at 4°C. Secondary fixation was done in 1% osmium tetroxide. Fixed cells were dehydrated in various grades of acetone followed by clearing with toluene and infiltration with decreasing toluene: resin ratios. Blocks were prepared in epoxy resin. From each block 3 ultrathin sections were mounted onto copper grids and viewed on high resolution transmission electron microscope, TECNAI 200 Kv transmission electron microscopy (Fei; Electron Optics, Hillsboro, OR). At least 5–10 random areas were chosen for viewing from each section.
Immunocytochemistry for Ki67
Control and Dox-treated cells grown on coverslips were serum stimulated for 24 hours followed by fixation in 4% formaldehyde. Cells were blocked and permeabilized in 3% bovine serum albumin containing 0.1% Triton X-100 for 1 hour and incubated with Ki67 antibody overnight at 4°C. This in turn was detected by horseradish peroxidase–labeled secondary antibody using DAB (3,3′-diaminobenzidine) (Cat# QD420-YIKE; BioGenex, Fremont, CA) for color development.
Immunohistochemistry
Formalin fixed paraffin embedded tissue sections (4 μM) were deparaffinized in xylene and rehydrated in decreasing grades of ethanol. Tissue sections were blocked in 3% H2O2 for 10 minutes followed by antigen retrieval in either citrate buffer (pH 6) or Tris-EDTA buffer (pH 9). Power block was performed using BioGenex kit for 5 minutes. Slides were incubated with primary antibody overnight at 4°C followed by treatment with Super Enhancer (Cat# QD420; YIKE BioGenex, Fremont, CA) for 20 minutes. This was then detected by incubation with horseradish peroxidase–tagged antibody followed by color development using DAB as chromogen. Hepatocytes were distinguished based on size and polygonal shape, further the cord arrangement of the hepatocytes as evident in the images also helped in delineating them from the other cell types present in the portal and sinusoidal area. Stained slides were photographed in bright-field microscope (Nikon ECLIPSE Ni; Nikon Instruments Inc) and evaluated by a pathologist based on % positivity (positive cells/total cells × 100) and staining intensity (0 = absent, 1 = minimal, 2 = moderate, 3 = severe). IHC scores were calculated as % positivity × intensity.
Plasmid Constructs, Transfection, and Generation of Stable Cell Clones
Full length human CLPP cDNA was cloned in pEGFP-N1 at XhoI and BamHI sites. pEGFP-N1 empty backbone was used as control. GFP and CLPP-GFP plasmids were transfected in HepG2, Huh7 and PCH5CH8 cells using Lipofectamine LTX with Plus Reagent (Cat#15338100; Invitrogen). These cells were selected and cultured in presence of Geneticin (500 μg/mL) (Cat#10131027; Gibco, Carlsbad, CA). Stable cell lines of HepG2 and Huh7 were established by dilution plating. Percentage of GFP+ cells were evaluated by flow cytometry.
Mitochondrial DNA Estimation
To estimate mitochondrial DNA depletion, genomic DNA was isolated using phenol, chloroform, or isoamylalcohol. mtDNA copy number was determined by coamplifying mitochondria encoding 12S rRNA and nuclear encoding 18S rRNA as reference gene. SYBR green chemistry was used for qPCR amplification.
Mitochondria Isolation and Oxygen Consumption Rate
Mitochondria were isolated from 2 × 107 cultured cells using Mitochondria Isolation Kit (Cat #89874; Thermo Fisher Scientific) according to the manufacturer’s protocol. The isolated mitochondrial fraction was resuspended in suspension buffer (HEPES [20 mM, pH 7.2], sucrose [0.3M], EDTA [1 mM], MgCl2 [2 mM], KH2PO4 [0.5 mM]) and the change in dissolved oxygen concentration was monitored every 30 seconds by using TBR1025 single-channel free radical analyzer (World Precision Instruments, Sarasota, FL).
Mitochondrial Stress Test
OCR and extracellular acidification rate was analyzed using Seahorse XF24 Analyzer and Seahorse XF Cell Mito Stress Test Kit (Cat#103015; Agilent/Seahorse Bioscience, Santa Clara, CA). FCCP concentration and cell density was standardized before experiment setup. Sensor cartridge was hydrated and equal numbers of cells were seeded onto Seahorse XF24 Cell Culture Microplates 1 day before assay. On the day of assay the cartridge was loaded with oligomycin (10 μM), FCCP (5 μM for HepG2 and 2.5 μM for Huh7) and rotenone/antimycin A (5 μM). The compounds were serially injected to measure the basal and the changes in OCR after addition of the electron transport chain inhibitors. After the end of the assay, total protein content in each well was quantified by BCA Protein Assay Kit (Cat#23225; Thermo Scientific) and was used to normalize the OCR values.
Statistics
All statistical analysis were done using GraphPad Prism version 6.01 for Windows (GraphPad Software, San Diego, CA). Two tailed Student’s t test was used for cell culture experiments to arrive at the P values between 2 different conditions. For clinical samples, Kruskal Wallis 1-way analysis of variance with Dunn’s multiple comparison tests was used to calculate P values between the groups. Differences were considered statistically significant with P values <.05. Data were represented as mean ± SD from biological replicates.
Acknowledgments
The authors thank Professor T Ramasarma and Dr. Lucy Anderson for critical reading and editing of the manuscript. BS is a recipient of UGC senior research fellowship. The authors thank Dr. Nobuyuki Kato, Okayama University, for providing us the PH5CH8 cells. They also thank DST-FIST for infrastructural support. They also thank the anonymous reviewers for their valuable inputs.
Footnotes
Author contributions BS: Performed all experiments, compiled and analysed data, and inputs in manuscript writing; GR: conceived and designed the study, analysed data and wrote manuscript; RN: helped in experimental protocols; AR: Read pathology slides and gave clinical inputs; NT: helped in FACS and provided research inputs; VP and SMS: provided clinical samples, diagnosis and inputs; SKS: necessary clinical inputs and manuscript editing; KJG and SP: helped in mitochondrial respiration assay using Clarke’s electrode.
Conflicts of interest The authors disclose no conflicts.
References
- 1.Tsochatzis E.A., Bosch J., Burroughs A.K. Liver cirrhosis. Lancet. 2014;383:1749–1761. doi: 10.1016/S0140-6736(14)60121-5. [DOI] [PubMed] [Google Scholar]
- 2.Aravinthan A., Pietrosi G., Hoare M., Jupp J., Marshall A., Verrill C., Davies S., Bateman A., Sheron N., Allison M., Alexander G.J. Hepatocyte expression of the senescence marker p21 is linked to fibrosis and an adverse liverrelated outcome in alcohol-related liver disease. PLoS One. 2013;8:e72904. doi: 10.1371/journal.pone.0072904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paradis V., Youssef N., Dargere D., Ba N., Bonvoust F., Deschatrette J., Bedossa P. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Human Pathol. 2001;32:327–332. doi: 10.1053/hupa.2001.22747. [DOI] [PubMed] [Google Scholar]
- 4.Aravinthan A., Scarpini C., Tachtatzis P., Verma S., Penrhyn-Lowe S., Harvey R., Davies S.E., Allison M., Coleman N., Alexander G. Hepatocyte senescence predicts progression in non-alcohol-related fatty liver disease. J Hepatol. 2013;58:549–556. doi: 10.1016/j.jhep.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 5.Dooley S., ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoare M., Das T., Alexander G. Ageing, telomeres, senescence, and liver injury. J Hepatol. 2010;53:950–961. doi: 10.1016/j.jhep.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H., Ju Z., Rudolph K.L. Telomere shortening and ageing. Z Gerontol Geriatr. 2007;40:314–324. doi: 10.1007/s00391-007-0480-0. [DOI] [PubMed] [Google Scholar]
- 8.Natarajan S.K., Thomas S., Ramamoorthy P., Basivireddy J., Pulimood A.B., Ramachandran A., Balasubramanian K.A. Oxidative stress in the development of liver cirrhosis: a comparison of 2 different experimental models. J Gastroenterol Hepatol. 2006;21:947–957. doi: 10.1111/j.1440-1746.2006.04231.x. [DOI] [PubMed] [Google Scholar]
- 9.Reyes-Gordillo K., Shah R., Muriel P. Oxidative Stress and inflammation in hepatic diseases: current and future therapy. Oxid Med Cell Longev. 2017;2017:3140673. doi: 10.1155/2017/3140673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimura T., Sasatani M., Kamiya K., Kawai H., Inaba Y., Kunugita N. Mitochondrial reactive oxygen species perturb AKT/cyclin D1 cell cycle signaling via oxidative inactivation of PP2A in lowdose irradiated human fibroblasts. Oncotarget. 2016;7:3559–3570. doi: 10.18632/oncotarget.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grattagliano I., Russmann S., Diogo C., Bonfrate L., Oliveira P.J., Wang D.Q., Portincasa P. Mitochondria in chronic liver disease. Curr Drug Targets. 2011;12:879–893. doi: 10.2174/138945011795528877. [DOI] [PubMed] [Google Scholar]
- 12.Hoshino A., Mita Y., Okawa Y., Ariyoshi M., Iwai-Kanai E., Ueyama T., Ikeda K., Ogata T., Matoba S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 13.Pang L., Liu K., Liu D., Lv F., Zang Y., Xie F., Yin J., Shi Y., Wang Y., Chen D. Differential effects of reticulophagy and mitophagy on nonalcoholic fatty liver disease. Cell Death Dis. 2018;9:90. doi: 10.1038/s41419-017-0136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams J.A., Ding W.X. A Mechanistic review of mitophagy and its role in protection against alcoholic liver disease. Biomolecules. 2015;5:2619–2642. doi: 10.3390/biom5042619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shpilka T., Haynes C.M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 16.Schulz A.M., Haynes C.M. UPR(mt)-mediated cytoprotection and organismal aging. Biochim Biophys Acta. 2015;1847:1448–1456. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gariani K., Menzies K.J., Ryu D., Wegner C.J., Wang X., Ropelle E.R., Moullan N., Zhang H., Perino A., Lemos V., Kim B., Park Y.K., Piersigilli A., Pham T.X., Yang Y., Ku C.S., Koo S.I., Fomitchova A., Canto C., Schoonjans K., Sauve A.A., Lee J.Y., Auwerx J. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology. 2016;63:1190–1204. doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhaskaran S., Pharaoh G., Ranjit R., Murphy A., Matsuzaki S., Nair B.C., Forbes B., Gispert S., Auburger G., Humphries K.M., Kinter M., Griffin T.M., Deepa S.S. Loss of mitochondrial protease ClpP protects mice from diet-induced obesity and insulin resistance. EMBO Rep. 2018;19:e45009. doi: 10.15252/embr.201745009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker C., Kukat A., Szczepanowska K., Hermans S., Senft K., Brandscheid C.P., Maiti P., Trifunovic A. CLPP deficiency protects against metabolic syndrome but hinders adaptive thermogenesis. EMBO Rep. 2018;19:e45126. doi: 10.15252/embr.201745126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anwar T., Khosla S., Ramakrishna G. Increased expression of SIRT2 is a novel marker of cellular senescence and is dependent on wild type p53 status. Cell Cycle. 2016;15:1883–1897. doi: 10.1080/15384101.2016.1189041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato N., Ikeda M., Sugiyama K., Mizutani T., Tanaka T., Shimotohno K. Hepatitis C virus population dynamics in human lymphocytes and hepatocytes infected in vitro. J Gen Virol. 1998;79:1859–1869. doi: 10.1099/0022-1317-79-8-1859. [DOI] [PubMed] [Google Scholar]
- 22.Bird T.G., Muller M., Boulter L., Vincent D.F., Ridgway R.A., Lopez-Guadamillas E., Lu W.Y., Jamieson T., Govaere O., Campbell A.D., Ferreira-Gonzalez S., Cole A.M., Hay T., Simpson K.J., Clark W., Hedley A., Clarke M., Gentaz P., Nixon C., Bryce S., Kiourtis C., Sprangers J., Nibbs R.J.B., Van Rooijen N., Bartholin L., McGreal S.R., Apte U., Barry S.T., Iredale J.P., Clarke A.R., Serrano M., Roskams T.A., Sansom O.J., Forbes S.J. TGFbeta inhibition restores a regenerative response in acute liver injury by suppressing paracrine senescence. Sci Transl Med. 2018;10:eaan1230. doi: 10.1126/scitranslmed.aan1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon Y.H., Jovanovic A., Serfas M.S., Kiyokawa H., Tyner A.L. p21 functions to maintain quiescence of p27-deficient hepatocytes. J Biol Chem. 2002;277:41417–41422. doi: 10.1074/jbc.M203388200. [DOI] [PubMed] [Google Scholar]
- 24.Inoue Y., Tomiya T., Nishikawa T., Ohtomo N., Tanoue Y., Ikeda H., Koike K. Induction of p53-dependent p21 limits proliferative activity of rat hepatocytes in the presence of hepatocyte growth factor. PLoS One. 2013;8:e78346. doi: 10.1371/journal.pone.0078346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwan R., Brady G.F., Brzozowski M., Weerasinghe S.V., Martin H., Park M.J., Brunt M.J., Menon R.K., Tong X., Yin L., Stewart C.L., Omary M.B. Hepatocyte-specific deletion of mouse lamin A/C leads to male-selective steatohepatitis. Cell Mol Gastroenterol Hepatol. 2017;4:365–383. doi: 10.1016/j.jcmgh.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrikx T., Schnabl B. Lamin deficiency in the liver sets the stage for nonalcoholic steatohepatitis development in males. Cell Mol Gastroenterol Hepatol. 2017;4:441–442. doi: 10.1016/j.jcmgh.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brady G.F., Kwan R., Bragazzi Cunha J., Elenbaas J.S., Omary M.B. Lamins and lamin-associated proteins in gastrointestinal health and disease. Gastroenterology. 2018;154:1602–1619.e1. doi: 10.1053/j.gastro.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L., Yannam G.R., Nishikawa T., Yamamoto T., Basma H., Ito R., Nagaya M., Dutta-Moscato J., Stolz D.B., Duan F., Kaestner K.H., Vodovotz Y., Soto-Gutierrez A., Fox I.J. The microenvironment in hepatocyte regeneration and function in rats with advanced cirrhosis. Hepatology. 2012;55:1529–1539. doi: 10.1002/hep.24815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravinthan A.D., Alexander G.J.M. Senescence in chronic liver disease: is the future in aging? J Hepatol. 2016;65:825–834. doi: 10.1016/j.jhep.2016.05.030. [DOI] [PubMed] [Google Scholar]
- 31.Gispert S., Parganlija D., Klinkenberg M., Drose S., Wittig I., Mittelbronn M., Grzmil P., Koob S., Hamann A., Walter M., Buchel F., Adler T., Hrabe de Angelis M., Busch D.H., Zell A., Reichert A.S., Brandt U., Osiewacz H.D., Jendrach M., Auburger G. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum MOl Genet. 2013;22:4871–4887. doi: 10.1093/hmg/ddt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deepa S.S., Bhaskaran S., Ranjit R., Qaisar R., Nair B.C., Liu Y., Walsh M.E., Fok W.C., Van Remmen H. Down-regulation of the mitochondrial matrix peptidase ClpP in muscle cells causes mitochondrial dysfunction and decreases cell proliferation. Free Radic Biol Med. 2016;91:281–292. doi: 10.1016/j.freeradbiomed.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczepanowska K., Maiti P., Kukat A., Hofsetz E., Nolte H., Senft K., Becker C., Ruzzenente B., Hornig-Do H.T., Wibom R., Wiesner R.J., Kruger M., Trifunovic A. CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J. 2016;35:2566–2583. doi: 10.15252/embj.201694253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pryde K.R., Taanman J.W., Schapira A.H. A LON-ClpP proteolytic axis degrades complex I to extinguish ROS production in depolarized mitochondria. Cell Rep. 2016;17:2522–2531. doi: 10.1016/j.celrep.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole A., Wang Z., Coyaud E., Voisin V., Gronda M., Jitkova Y., Mattson R., Hurren R., Babovic S., Maclean N., Restall I., Wang X., Jeyaraju D.V., Sukhai M.A., Prabha S., Bashir S., Ramakrishnan A., Leung E., Qia Y.H., Zhang N., Combes K.R., Ketela T., Lin F., Houry W.A., Aman A., Al-Awar R., Zheng W., Wienholds E., Xu C.J., Dick J., Wang J.C., Moffat J., Minden M.D., Eaves C.J., Bader G.D., Hao Z., Kornblau S.M., Raught B., Schimmer A.D. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2015;27:864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seiferling D., Szczepanowska K., Becker C., Senft K., Hermans S., Maiti P., Konig T., Kukat A., Trifunovic A. Loss of CLPP alleviates mitochondrial cardiomyopathy without affecting the mammalian UPRmt. EMBO Rep. 2016;17:953–964. doi: 10.15252/embr.201642077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thuluvath P.J., Kantsevoy S., Thuluvath A.J., Savva Y. Is cryptogenic cirrhosis different from NASH cirrhosis? J Hepatol. 2018;68:519–525. doi: 10.1016/j.jhep.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 38.de Franchis R. Evolving consensus in portal hypertension. Report of the Baveno IV consensus workshop on methodology of diagnosis and therapy in portal hypertension. J Hepatol. 2005;43:167–176. doi: 10.1016/j.jhep.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Rastogi A., Maiwall R., Bihari C., Ahuja A., Kumar A., Singh T., Wani Z.A., Sarin S.K. Cirrhosis histology and Laennec staging system correlate with high portal pressure. Histopathology. 2013;62:731–741. doi: 10.1111/his.12070. [DOI] [PubMed] [Google Scholar]
- 40.Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Peacocke M., Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9567. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]