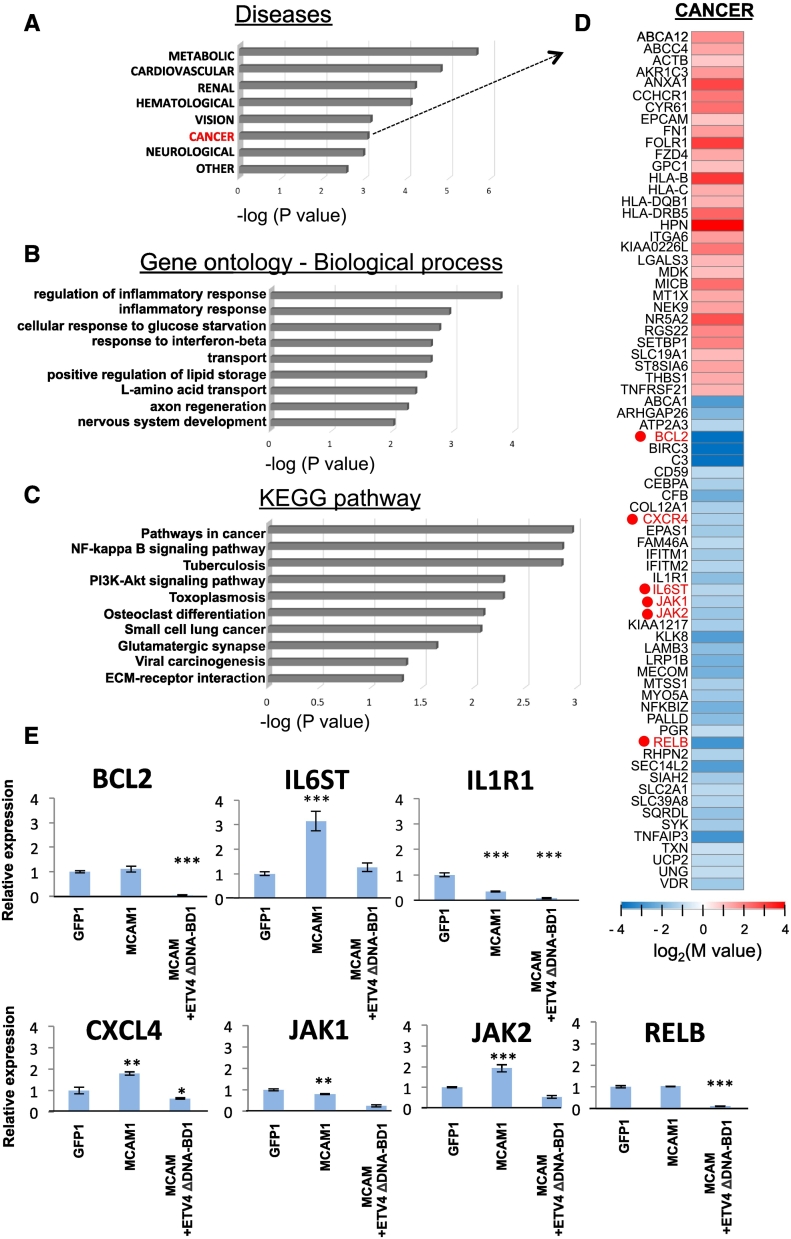
Figure 6.
RNA-seq analysis. A, Functional enrichment analysis (P < .05) was performed for protein-coding RNAs in GAD_DISEASE_CLASS with upregulated and downregulated genes in the MCF-7-derived MCAM+ETV4 ΔDNA-BD #1 clone in comparison to those in the MCAM #1 clone. B and C, For the downregulated genes, functional enrichment analysis (P < .05) was also performed in GO (B) and KEGG (C) analysis. D, Heat maps are displayed for the selected genes that are upregulated and downregulated in the cancer category of the disease clustering as indicated in (A). These analyses were performed using the database for annotation, visualization and integrated discovery (DAVID) v6.8 (http://david.ncifcr.gov/). E, Quantitative real-time PCR analysis was carried out in the indicated cells for the genes shown in (D). The relative expression level of each sample is shown after calibration with the TBP (a suitable housekeeping gene) value. Data are means ± SD, *P < .05, **P < .01 and ***P < .001. ND: not detected.