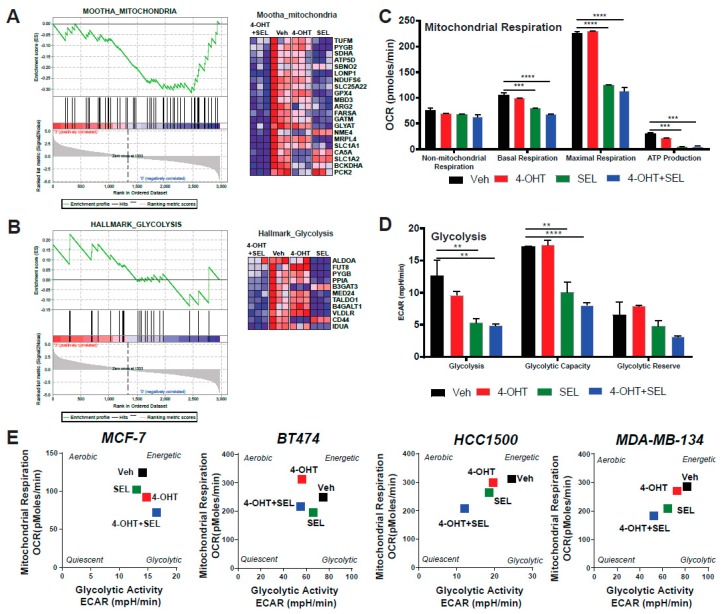
Figure 4.
ERα-XPO1 targeting changes the metabolic phenotype of breast cancer cells from an energetic to a quiescent profile. GSEA analysis of RNA-Seq data depicting the regulation of glycolytic (A) and mitochondrial respiration (B) pathways by the 4-OHT+SEL combination. ERα-XPO1 targeting decreased the glycolytic activity of BT474 cells. Glycolytic functions were determined by the glycolysis stress test. A two-way analysis of variance (ANOVA) model was used for statistical significance of drug treatments alone or in combination and values were presented as mean ± SEM from two independent experiments and all experiments were performed in triplicates. (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). (C,D) ERα-XPO1 targeting decreased the mitochondrial activity of BT474 cells. Mitochondrial activity parameters were measured by the mitochondrial stress test. A two-way analysis of variance (ANOVA) model was used for statistical significance calculations and values were presented as mean ± SEM from three independent experiments (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). Some key parameters of mitochondrial function (Basal respiration, proton leak, maximal respiration, spare respiratory capacity, non-mitochondrial respiration, ATP production, coupling efficiency and spare respiratory capacity) were calculated and presented for each treatment conditions. (E) Cell phenotype assay resulted in MCF-7, BT474, MDA-MB-134 and HCC1500 cells, which were treated with 4-OHT (10−6 M) and SEL (10−7) M for 24 h alone, for 24 h alone and in combination. Changes in metabolic profile was determined by measuring oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) under basal and stressed conditions with the cell phenotype assay. Values are presented as ± SEM from three independent experiments.