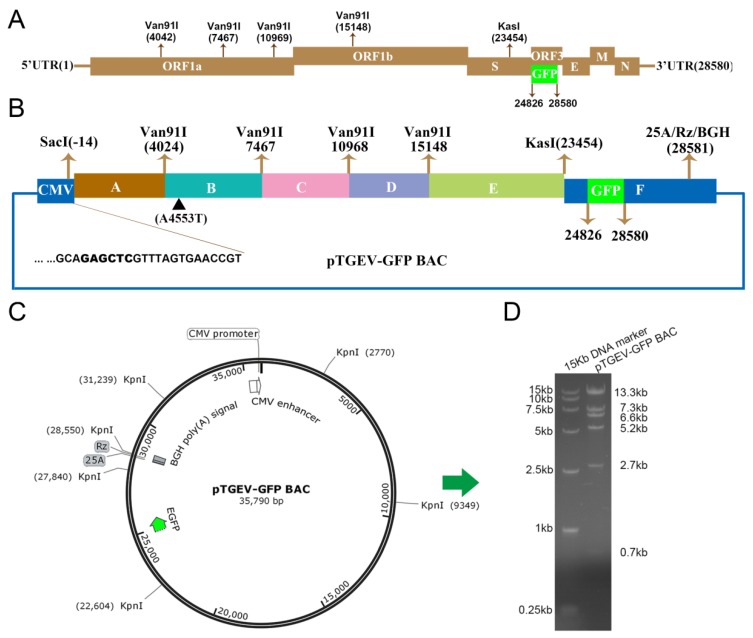
Figure 1.
Construction of the TGEV-GFP infectious bacterial artificial chromosome (BAC) clone. The number represents the nucleotide (nt) position in the TGEV genome. (A) Structure of the TGEV genome. The 5′ and 3′ UTRs represent the 5′ and 3′ untranslated regions, respectively. (B) The TGEV genome was divided into six contiguous cDNAs (A to F): A, −14 to 4024; B, 4025 to 7467; C, 7468 to 10,968; D, 10,969 to 15,148; E, 15,149 to 23,454; and F, 23,455 to 28,580. A4553T was introduced to ablate a natural Van91I site at nt 4553 (▲). The CMV promoter, fragment F, a 25-bp poly(A) tail (25A), hepatitis delta virus self-cleaving ribozyme sequence, Rz (HDV RZ), and the bovine growth hormone (BGH) transcriptional terminal signal were inserted into the BAC to form the final recipient BAC vector or subclone F. The following restriction sites are noted: SacI (−14), Van91I (4024, 7467, 10,968, and 15,148) and KasI (23,454). The EGFP gene replacing open reading frame 3 (ORF3) is noted at 24,826 and 25,692. (C) Schematic map of pTGEV-GFP BAC restriction enzyme digestion by KpnI. (D) The left lane is the DL15000 DNA marker, and the right lane is the product of pTGEV-GFP BAC by KpnI digestion.