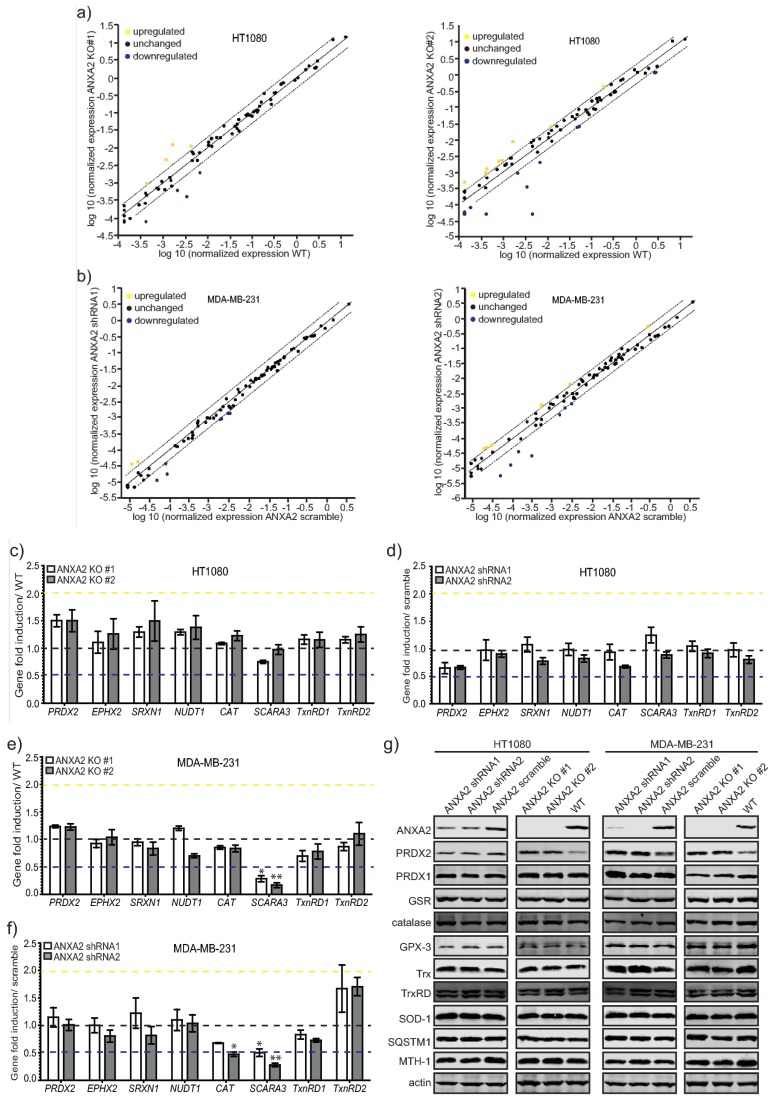
Figure 5.
Analysis of ROS related genes and proteins in ANXA2 depleted versus control cancer cells. (a) HT1080 ANXA2 KO #1; ANXA2 KO #2 or WT or (b) MDA-MB-231 ANXA2 shRNA1; ANXA2 shRNA2 or ANXA2 scramble cells were plated in 100 mm plates for 48 h. After what RNA extraction was performed using the RNeasy mini kit (Qiagen, Manchester, UK) according to the manufacturer′s instructions. A panel of 86 ROS dependent genes was analysed using the RT2 Profiler™ PCR Array Human Oxidative Stress (Qiagen, Manchester, UK) according to the manufacturer’s instructions in a LightCycler 96 instrument (Roche, Basel, Switzerland). (c) HT1080 ANXA2 KO #1; ANXA2 KO #2 or WT; (d) HT1080 ANXA2 shRNA1; ANXA2 shRNA2 or ANXA2 scramble; (e) MDA-MB-231 ANXA2 KO #1; ANXA2 KO #2 or WT; (f) MDA-MB-231 ANXA2 shRNA1; ANXA2 shRNA2 or ANXA2 scramble cells were plated in 100 mm plates for 48 h. RNA extraction was performed using the RNeasy mini kit (Qiagen, Manchester, UK) according to the manufacturer´s instructions. The gene expression was determined by qRT-PCR using the One-step NZYSpeedy RT-qPCR Green kit (Nzytech, Lisbon, Portugal) according to manufacturer´s instructions. The reaction was carried out using the Lightcycler 96 instrument (Roche, Basel, Switzerland). Gene expression levels were normalized to RPLP0 mRNA using the 2−ΔΔCT method (Livak KJ, Schmittgen TD). Error bars represent the standard deviation obtained from three independent experiments. Statistical analysis was evaluated using two-tailed Student’s t-test. In every case a p-value of less than 0.05 (*), less than 0.01(**) and 0.001 (***) was considered statistically significant. (g) HT1080 ANXA2 shRNA1, ANXA2 shRNA2 and scramble (first panel); HT1080 ANXA2 KO #1, ANXA2 KO #2 and WT (second panel); MDA-MB-231 ANXA2 shRNA1, ANXA2 shRNA2 and scramble (third panel); MDA-MB-231 ANXA2 KO #1, ANXA2 KO #2 and WT (fourth panel) cells were lysed and 20 µg of each protein extract was subjected to SDS-PAGE, transferred onto nitrocellulose membranes and analyzed by western blotting with the antibodies indicated. Results are representative of three independent experiments (n = 3).