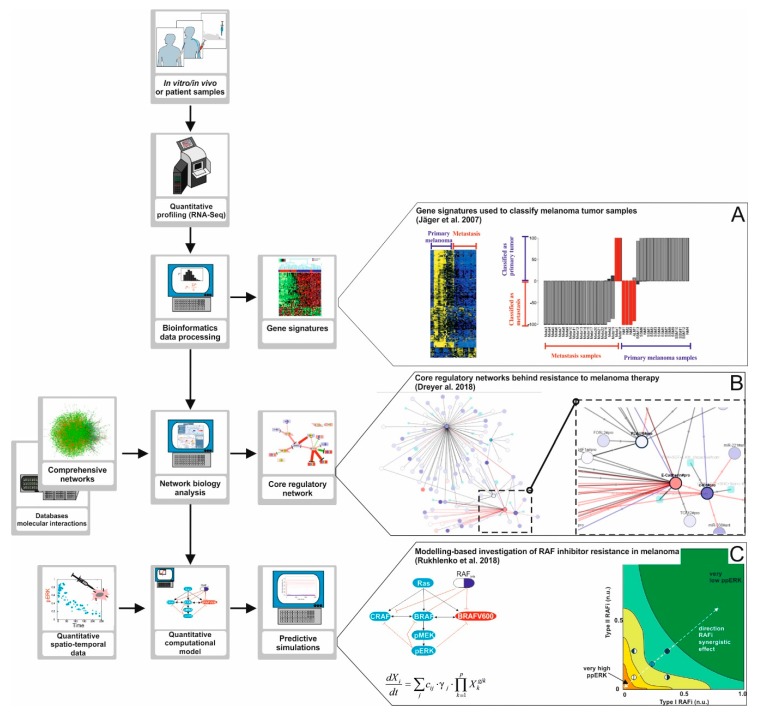
Figure 3.
A road map to apply Systems Biology approaches in melanoma research. The development of high-throughput data generation technologies like next-generation sequencing, proteomics, and phosphoproteomics opens opportunities to investigate the mechanisms of emergence for BRAF inhibitor therapy resistance in melanoma in depth. (A) High-throughput data analysis. The data obtained from the quantitative profiling of tumor samples can be processed and analyzed making use of bioinformatics pipelines and algorithms. Using this approach, one can discover gene signatures, that is, ensembles of genes collectively deregulated in tumor samples from a group of patients. In the study of Jäger and co-workers [62], tumor samples were profiled and the data obtained was used to find a predictive gene signature for metastatic melanoma. (B) Network Biology. The processed high-throughput data can also be analyzed using network biology approaches. In this case, the data are mapped into large molecular networks, and computational algorithms are used to detect core regulatory networks, highly interconnected fractions of larger networks that are differentially regulated in, for example, treatment responders versus non-responders. The core network, compared to a gene signature, gives additional information because it helps constructing hypotheses about the molecular mechanisms linking the differentially regulated genes. In a study by Dreyer and co-workers [66] we reconstructed a comprehensive network accounting for signaling pathways commonly deregulated in melanoma. The network was used to detect a core regulatory network discriminating responders from non-responders in a cohort or patients treated with anti-PD-1 immunotherapy. The comprehensive network derived is available at www.vcells.net/melanoma. (C) Computational modelling of regulatory pathways. High- throughput data, core regulatory networks, and additional quantitative data like drug dose response curves or time series of signal pathway activation can be used to derive, characterize, and train computational models of regulatory networks relevant for therapy. Computational simulations performed with these models can be used to formulate hypotheses, design experiments, or even to (re)design combinatory therapies. In a study by Rukhlenko and co-workers [67] simulations of a detailed computational model of the RAF/MEK/ERK signaling pathway in melanoma were used to predict combinatory BRAF inhibitor therapy able to re-sensitize therapy resistant melanoma cell lines.